|
|
|
|

|
| Sample: |
DNA-directed RNA polymerase subunit alpha dimer, 78 kDa Mycobacterium tuberculosis protein
DNA-directed RNA polymerase subunit beta monomer, 129 kDa Mycobacterium tuberculosis protein
DNA-directed RNA polymerase subunit beta' monomer, 147 kDa Mycobacterium tuberculosis protein
DNA-directed RNA polymerase subunit omega monomer, 12 kDa Mycobacterium tuberculosis protein
|
| Buffer: |
10 mM Tris-Cl, 100 mM NaCl, 10 mM MgCl2, 0.1 mM EDTA, 5% Glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Mar 8
|
Structural and biochemical characterization of RNA polymerase core and UvrD complex: a key component in transcription coupled DNA repair
Ravishankar Ramachandran
|
| RgGuinier |
5.1 |
nm |
| Dmax |
19.3 |
nm |
| VolumePorod |
262 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
DNA-directed RNA polymerase subunit alpha dimer, 75 kDa Mycobacterium tuberculosis protein
DNA-directed RNA polymerase subunit beta monomer, 129 kDa Mycobacterium tuberculosis protein
DNA-directed RNA polymerase subunit beta' monomer, 147 kDa Mycobacterium tuberculosis protein
DNA-directed RNA polymerase subunit omega monomer, 12 kDa Mycobacterium tuberculosis protein
ATP-dependent DNA helicase UvrD1 monomer, 85 kDa Mycobacterium tuberculosis protein
|
| Buffer: |
10 mM Tris-Cl, 100 mM NaCl, 10 mM MgCl2, 0.1 mM EDTA, 5% Glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Mar 11
|
Structural and biochemical characterization of RNA polymerase core and UvrD complex: a key component in transcription coupled DNA repair
Ravishankar Ramachandran
|
| RgGuinier |
7.5 |
nm |
| Dmax |
22.2 |
nm |
| VolumePorod |
898 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Iron-sulfur cluster assembly protein trimer, 42 kDa Plasmodium falciparum (isolate … protein
|
| Buffer: |
50 mM Tris-Cl, 300 mM NaCl, 5% Glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR-Central Drug Research Institute on 2019 May 5
|
[Fe-S] cluster biogenesis and unusual assembly of the ISC scaffold complex in Plasmodium falciparum mitochondrion
Ravishankar Ramachandran
|
| RgGuinier |
5.4 |
nm |
| Dmax |
11.1 |
nm |
| VolumePorod |
177 |
nm3 |
|
|
|
|
|
|
|
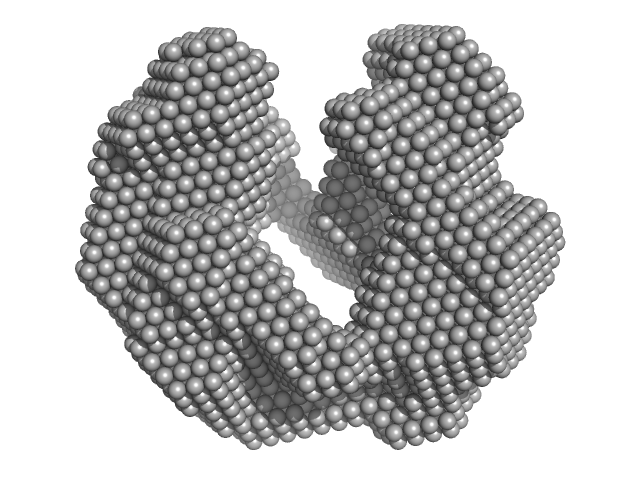
| Sample: |
Iron-sulfur cluster assembly protein tetramer, 56 kDa Plasmodium falciparum (isolate … protein
Cysteine desulfurase, putative tetramer, 204 kDa Plasmodium falciparum (isolate … protein
|
| Buffer: |
50 mM Tris-Cl, 300 mM NaCl, 5% Glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR-Central Drug Research Institute on 2019 Sep 4
|
[Fe-S] cluster biogenesis and unusual assembly of the ISC scaffold complex in Plasmodium falciparum mitochondrion
Ravishankar Ramachandran
|
| RgGuinier |
6.4 |
nm |
| Dmax |
17.6 |
nm |
| VolumePorod |
1100 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Cysteine desulfurase, putative tetramer, 204 kDa Plasmodium falciparum (isolate … protein
Iron-sulfur cluster assembly protein tetramer, 56 kDa Plasmodium falciparum (isolate … protein
Protein ISD11 tetramer, 43 kDa Plasmodium falciparum (isolate … protein
|
| Buffer: |
50 mM Tris-Cl, 300 mM NaCl, 5% Glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR-Central Drug Research Institute on 2019 Sep 19
|
[Fe-S] cluster biogenesis and unusual assembly of the ISC scaffold complex in Plasmodium falciparum mitochondrion
Ravishankar Ramachandran
|
| RgGuinier |
7.1 |
nm |
| Dmax |
17.9 |
nm |
| VolumePorod |
950 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Putative DNA binding protein dimer, 37 kDa Bacillus subtilis subsp. … protein
|
| Buffer: |
20 mM HEPES 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR - Institute of Microbial Technology (IMTech) on 2016 Aug 17
|
YeeF, a polymorphic toxin from Bacillus subtilis, is a ribosomal RNase with promiscuous DNA binding property
Krishan Gopal
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
55 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
YezG, cognate immunity protein of YeeF monomer, 19 kDa Bacillus subtilis subsp. … protein
|
| Buffer: |
20 mM HEPES 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR - Institute of Microbial Technology (IMTech) on 2016 Aug 17
|
YeeF, a polymorphic toxin from Bacillus subtilis, is a ribosomal RNase with promiscuous DNA binding property
Krishan Gopal
|
| RgGuinier |
2.1 |
nm |
| Dmax |
6.1 |
nm |
| VolumePorod |
46 |
nm3 |
|
|
|
|
|
|

|
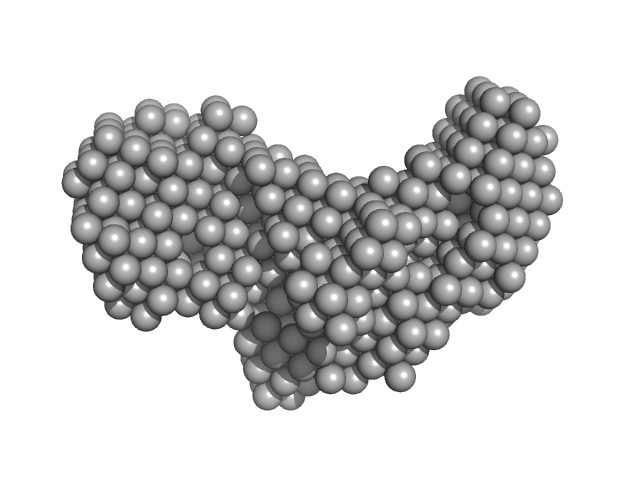
| Sample: |
Putative DNA binding protein dimer, 37 kDa Bacillus subtilis subsp. … protein
YezG, cognate immunity protein of YeeF monomer, 19 kDa Bacillus subtilis subsp. … protein
|
| Buffer: |
20 mM HEPES 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR - Institute of Microbial Technology (IMTech) on 2016 Aug 18
|
YeeF, a polymorphic toxin from Bacillus subtilis, is a ribosomal RNase with promiscuous DNA binding property
Krishan Gopal
|
| RgGuinier |
4.0 |
nm |
| Dmax |
14.1 |
nm |
| VolumePorod |
115 |
nm3 |
|
|
|
|
|
|
|
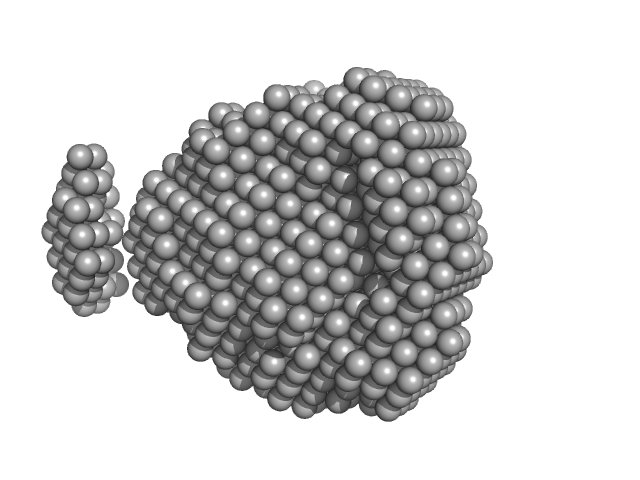
| Sample: |
Heat shock 70 kDa protein 1A monomer, 70 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris HCl, 50 mM NaCl, 5 mM Sodium Phosphate, 5 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS1 Beamline, Brazilian Synchrotron Light Laboratory on 2018 May 18
|
Structural, thermodynamic and functional studies of human 71 kDa heat shock cognate protein (HSPA8/hHsc70)
Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics :140719 (2021)
Silva N, de Camargo Rodrigues L, Dores-Silva P, Montanari C, Ramos C, Barbosa L, Borges J
|
| RgGuinier |
3.7 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
138 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Fructokinase, PfkB monomer, 32 kDa Mycobacterium marinum (strain … protein
|
| Buffer: |
20 mM Tris-HCl, 100 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2019 Dec 17
|
Structural analysis and functional study of phosphofructokinase B (PfkB) from Mycobacterium marinum
Biochemical and Biophysical Research Communications (2021)
Gao B, Ji R, Li Z, Su X, Li H, Sun Y, Ji C, Gan J, Li J
|
| RgGuinier |
2.0 |
nm |
| Dmax |
6.6 |
nm |
| VolumePorod |
58 |
nm3 |
|
|