|
|
|
|

|
| Sample: |
Extracelllular iron oxide respiratory system periplasmic decaheme cytochrome c component MtrA monomer, 39 kDa Shewanella oneidensis (strain … protein
Extracellular iron oxide respiratory system surface decaheme cytochrome c component MtrC monomer, 77 kDa Shewanella oneidensis (strain … protein
Extracellular iron oxide respiratory system outer membrane component MtrB monomer, 75 kDa Shewanella oneidensis (strain … protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 2.8 mM Fos-choline 12, 13% D2O, pH: 7.8 |
| Experiment: |
SANS
data collected at D22, Institut Laue-Langevin (ILL) on 2020 Jan 13
|
Bespoke Biomolecular Wires for Transmembrane Electron Transfer: Spontaneous Assembly of a Functionalized Multiheme Electron Conduit
Frontiers in Microbiology 12 (2021)
Piper S, Edwards M, van Wonderen J, Casadevall C, Martel A, Jeuken L, Reisner E, Clarke T, Butt J
|
| RgGuinier |
4.7 |
nm |
| Dmax |
16.6 |
nm |
| VolumePorod |
154 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Extracelllular iron oxide respiratory system periplasmic decaheme cytochrome c component MtrA monomer, 39 kDa Shewanella oneidensis (strain … protein
Extracellular iron oxide respiratory system outer membrane component MtrB monomer, 75 kDa Shewanella oneidensis (strain … protein
Extracellular iron oxide respiratory system surface decaheme cytochrome c component MtrC monomer, 75 kDa Shewanella oneidensis (strain … protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 2.8 mM Fos-choline 12, 13% D2O, pH: 7.8 |
| Experiment: |
SANS
data collected at D22, Institut Laue-Langevin (ILL) on 2016 Sep 12
|
Bespoke Biomolecular Wires for Transmembrane Electron Transfer: Spontaneous Assembly of a Functionalized Multiheme Electron Conduit
Frontiers in Microbiology 12 (2021)
Piper S, Edwards M, van Wonderen J, Casadevall C, Martel A, Jeuken L, Reisner E, Clarke T, Butt J
|
| RgGuinier |
5.2 |
nm |
| Dmax |
17.1 |
nm |
| VolumePorod |
234 |
nm3 |
|
|
|
|
|
|

|
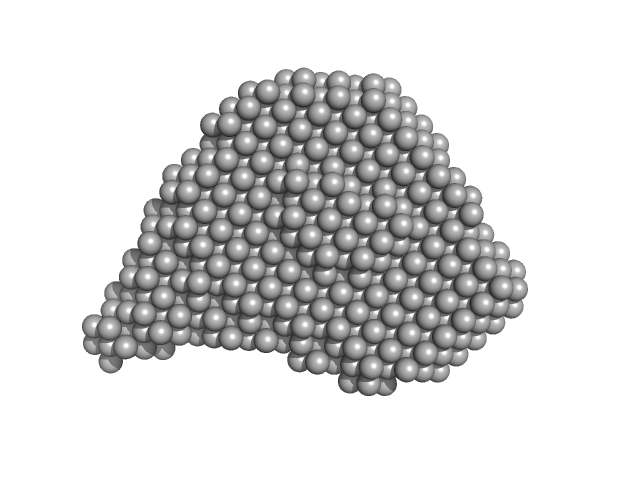
| Sample: |
Efa1/LifA protein monomer, 367 kDa Escherichia coli O127:H6 … protein
|
| Buffer: |
50 mM Bis-Tris, 100 mM NaCl, 3 % (v/v) glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Jun 29
|
Activity of lymphostatin, a lymphocyte inhibitory virulence factor of pathogenic Escherichia coli, is dependent on a cysteine protease motif.
J Mol Biol :167200 (2021)
Bease AG, Blackburn EA, Chintoan-Uta C, Webb S, Cassady-Cain RL, Stevens MP
|
| RgGuinier |
5.9 |
nm |
| Dmax |
19.3 |
nm |
| VolumePorod |
559 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Efa1/LifA protein monomer, 367 kDa Escherichia coli O127:H6 … protein
|
| Buffer: |
50 mM Bis-Tris, 100 mM NaCl, 3 % (v/v) glycerol, pH: 5.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Jun 29
|
Activity of lymphostatin, a lymphocyte inhibitory virulence factor of pathogenic Escherichia coli, is dependent on a cysteine protease motif.
J Mol Biol :167200 (2021)
Bease AG, Blackburn EA, Chintoan-Uta C, Webb S, Cassady-Cain RL, Stevens MP
|
| RgGuinier |
5.9 |
nm |
| Dmax |
19.7 |
nm |
| VolumePorod |
565 |
nm3 |
|
|
|
|
|
|

|
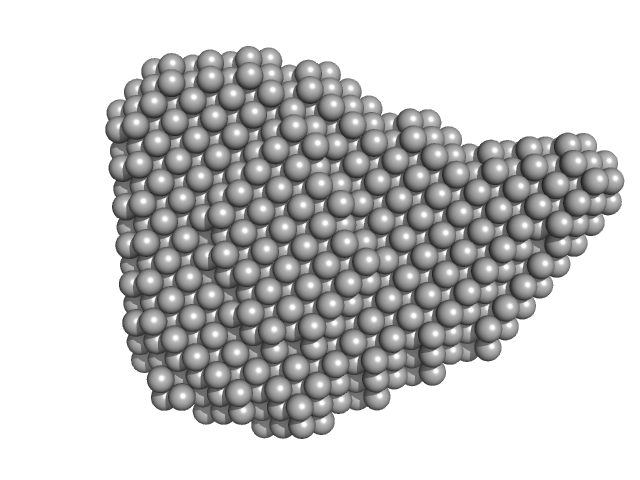
| Sample: |
Efa1/LifA protein (C1480A mutant) monomer, 367 kDa Escherichia coli O127:H6 … protein
|
| Buffer: |
50 mM Bis-Tris, 100 mM NaCl, 3 % (v/v) glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Jun 29
|
Activity of lymphostatin, a lymphocyte inhibitory virulence factor of pathogenic Escherichia coli, is dependent on a cysteine protease motif.
J Mol Biol :167200 (2021)
Bease AG, Blackburn EA, Chintoan-Uta C, Webb S, Cassady-Cain RL, Stevens MP
|
| RgGuinier |
5.9 |
nm |
| Dmax |
19.1 |
nm |
| VolumePorod |
557 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Efa1/LifA protein (C1480A mutant) monomer, 367 kDa Escherichia coli O127:H6 … protein
|
| Buffer: |
50 mM Bis-Tris, 100 mM NaCl, 3 % (v/v) glycerol, pH: 5.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Jun 29
|
Activity of lymphostatin, a lymphocyte inhibitory virulence factor of pathogenic Escherichia coli, is dependent on a cysteine protease motif.
J Mol Biol :167200 (2021)
Bease AG, Blackburn EA, Chintoan-Uta C, Webb S, Cassady-Cain RL, Stevens MP
|
| RgGuinier |
5.8 |
nm |
| Dmax |
19.3 |
nm |
| VolumePorod |
560 |
nm3 |
|
|
|
|
|
|

|
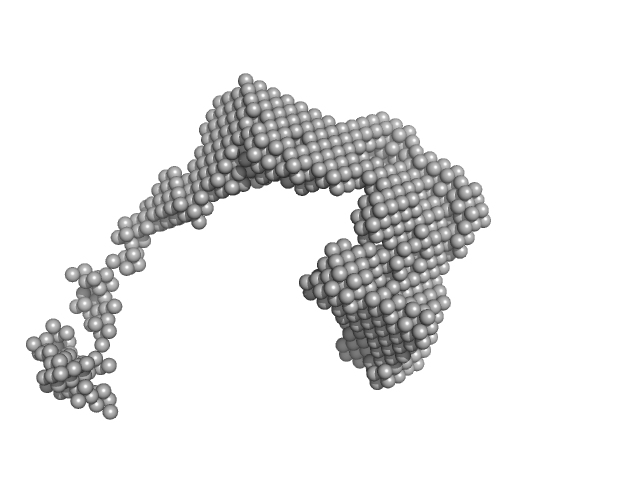
| Sample: |
Iota carbonic anhydrase tetramer, 260 kDa Thalassiosira pseudonana protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Mar 28
|
Structural Contour Map of the Iota Carbonic Anhydrase from the Diatom Thalassiosira pseudonana Using a Multiprong Approach.
Int J Mol Sci 22(16) (2021)
Jensen EL, Receveur-Brechot V, Hachemane M, Wils L, Barbier P, Parsiegla G, Gontero B, Launay H
|
| RgGuinier |
6.7 |
nm |
| Dmax |
25.0 |
nm |
| VolumePorod |
500 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Lysozyme amyloid fibril, 1 kDa Gallus gallus protein
|
| Buffer: |
0.2 M glycine-HCl, 80 mM NaCl, pH: 2.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Nov 29
|
Dependence of the Nanoscale Composite Morphology of Fe3O4 Nanoparticle-Infused Lysozyme Amyloid Fibrils on Timing of Infusion: A Combined SAXS and AFM Study
Molecules 26(16):4864 (2021)
Schroer M, Hu P, Tomasovicova N, Batkova M, Zakutanska K, Wu P, Kopcansky P
|
| RgGuinier |
38.4 |
nm |
| Dmax |
80.0 |
nm |
|
|
|
|
|
|
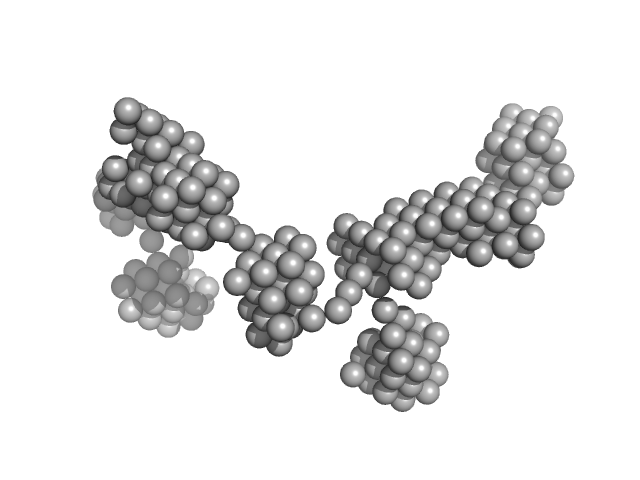
|
| Sample: |
Lysozyme amyloid fibril, 1 kDa Gallus gallus protein
Fe3O4 nanoparticles; nominal diameter 10 nm (hydrodynamic diameter) monomer, 1 kDa
|
| Buffer: |
0.2 M glycine-HCl, 80 mM NaCl, pH: 2.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Nov 29
|
Dependence of the Nanoscale Composite Morphology of Fe3O4 Nanoparticle-Infused Lysozyme Amyloid Fibrils on Timing of Infusion: A Combined SAXS and AFM Study
Molecules 26(16):4864 (2021)
Schroer M, Hu P, Tomasovicova N, Batkova M, Zakutanska K, Wu P, Kopcansky P
|
| RgGuinier |
22.3 |
nm |
| Dmax |
70.0 |
nm |
|
|
|
|
|
|

|
| Sample: |
Lysozyme amyloid fibril, 1 kDa Gallus gallus protein
Fe3O4 nanoparticles; nominal diameter 30 nm (hydrodynamic diameter) monomer, 1 kDa
|
| Buffer: |
0.2 M glycine-HCl, 80 mM NaCl, pH: 2.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Nov 29
|
Dependence of the Nanoscale Composite Morphology of Fe3O4 Nanoparticle-Infused Lysozyme Amyloid Fibrils on Timing of Infusion: A Combined SAXS and AFM Study
Molecules 26(16):4864 (2021)
Schroer M, Hu P, Tomasovicova N, Batkova M, Zakutanska K, Wu P, Kopcansky P
|
| RgGuinier |
22.5 |
nm |
| Dmax |
95.0 |
nm |
|
|