|
|
|
|
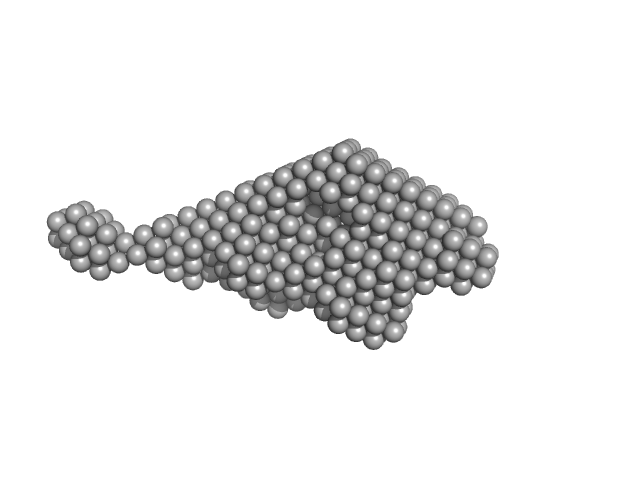
|
| Sample: |
Testican-2 monomer, 40 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES (Na), 130 mM NaCl, 5 % (v/v) glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Oct 18
|
The Central Region of Testican-2 Forms a Compact Core and Promotes Cell Migration
International Journal of Molecular Sciences 21(24):9413 (2020)
Krajnc A, Gaber A, Lenarčič B, Pavšič M
|
| RgGuinier |
3.4 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
82 |
nm3 |
|
|
|
|
|
|
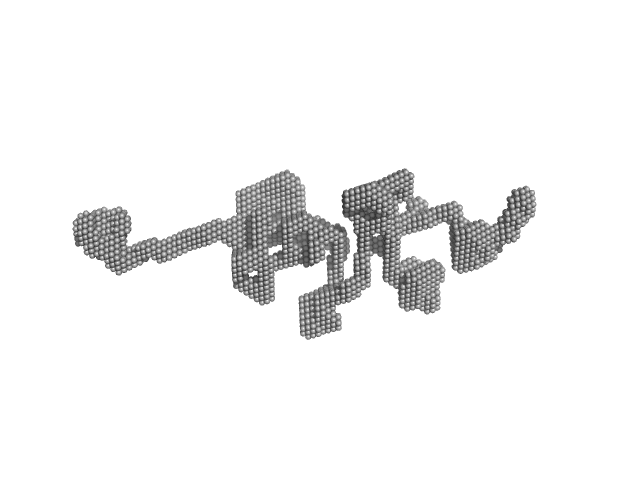
|
| Sample: |
Nucleolar RNA helicase 2 dimer, 182 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 500 mM NaCl, 10 % Glycerol, 2 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Nov 25
|
The Human RNA Helicase DDX21 Presents a Dimerization Interface Necessary for Helicase Activity
iScience 23(12):101811 (2020)
Marcaida M, Kauzlaric A, Duperrex A, Sülzle J, Moncrieffe M, Adebajo D, Manley S, Trono D, Dal Peraro M
|
| RgGuinier |
7.0 |
nm |
| Dmax |
31.9 |
nm |
| VolumePorod |
681 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Nucleolar RNA helicase 2 fragment 186-783 dimer, 138 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 500 mM NaCl, 10 % Glycerol, 2 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Mar 4
|
The Human RNA Helicase DDX21 Presents a Dimerization Interface Necessary for Helicase Activity
iScience 23(12):101811 (2020)
Marcaida M, Kauzlaric A, Duperrex A, Sülzle J, Moncrieffe M, Adebajo D, Manley S, Trono D, Dal Peraro M
|
| RgGuinier |
4.8 |
nm |
| Dmax |
17.6 |
nm |
| VolumePorod |
297 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Truncated HERV-K Rec response element monomer, 102 kDa Homo sapiens RNA
|
| Buffer: |
10mM Tris-HCl, 100mM KCl, and 0.5mM MgCl₂, pH: 8 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2018 Jul 17
|
Structural Mimicry Drives HIV-1 Rev-Mediated HERV-K Expression
Journal of Molecular Biology 432(24):166711 (2020)
O'Carroll I, Fan L, Kroupa T, McShane E, Theodore C, Yates E, Kondrup B, Ding J, Martin T, Rein A, Wang Y
|
| RgGuinier |
6.1 |
nm |
| Dmax |
21.8 |
nm |
| VolumePorod |
322 |
nm3 |
|
|
|
|
|
|
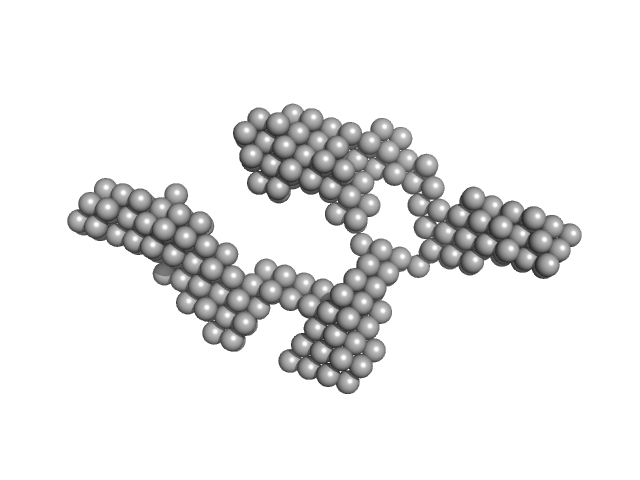
|
| Sample: |
TrΔSLIII HERV-K Rec response element monomer, 88 kDa Homo sapiens RNA
|
| Buffer: |
10mM Tris-HCl, 100mM KCl, and 0.5mM MgCl₂, pH: 8 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2018 Dec 7
|
Structural Mimicry Drives HIV-1 Rev-Mediated HERV-K Expression
Journal of Molecular Biology 432(24):166711 (2020)
O'Carroll I, Fan L, Kroupa T, McShane E, Theodore C, Yates E, Kondrup B, Ding J, Martin T, Rein A, Wang Y
|
| RgGuinier |
5.3 |
nm |
| Dmax |
19.2 |
nm |
| VolumePorod |
226 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
TrΔSLIIab HERV-K Rec response element monomer, 86 kDa Homo sapiens RNA
|
| Buffer: |
10mM Tris-HCl, 100mM KCl, and 0.5mM MgCl₂, pH: 8 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Feb 21
|
Structural Mimicry Drives HIV-1 Rev-Mediated HERV-K Expression
Journal of Molecular Biology 432(24):166711 (2020)
O'Carroll I, Fan L, Kroupa T, McShane E, Theodore C, Yates E, Kondrup B, Ding J, Martin T, Rein A, Wang Y
|
| RgGuinier |
5.0 |
nm |
| Dmax |
18.4 |
nm |
| VolumePorod |
201 |
nm3 |
|
|
|
|
|
|
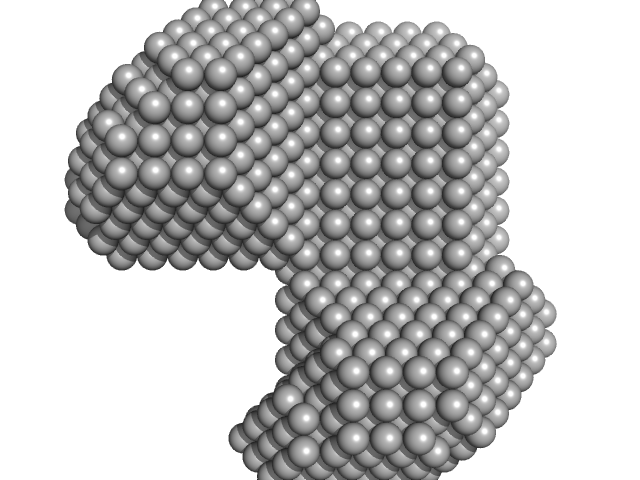
|
| Sample: |
LIM/homeobox protein Lhx3 monomer, 10 kDa Mus musculus protein
|
| Buffer: |
20 mM sodium phosphate monobasic/dibasic, 100 mM NaCl, 1 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2018 Nov 2
|
Contrasting DNA-binding behaviour by ISL1 and LHX3 underpins differential gene targeting in neuronal cell specification
Journal of Structural Biology: X :100043 (2020)
Smith N, Wilkinson-White L, Kwan A, Trewhella J, Matthews J
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
18 |
nm3 |
|
|
|
|
|
|
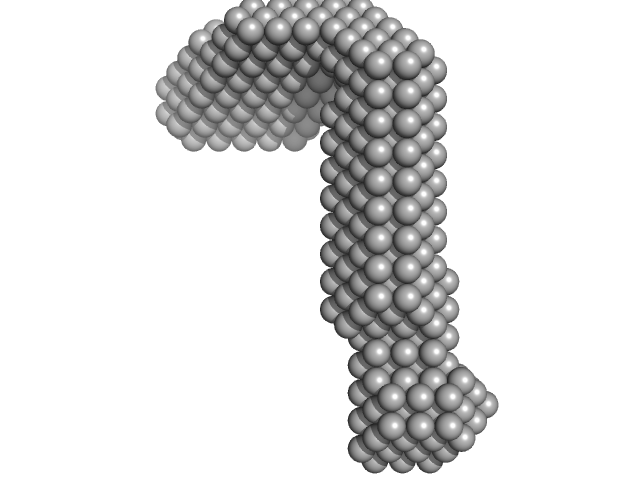
|
| Sample: |
LIM/homeobox protein Lhx3 monomer, 23 kDa Mus musculus protein
Insulin gene enhancer protein ISL-1 monomer, 4 kDa Mus musculus protein
|
| Buffer: |
20 mM sodium phosphate monobasic/dibasic, 100 mM NaCl, 1 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2018 Nov 2
|
Contrasting DNA-binding behaviour by ISL1 and LHX3 underpins differential gene targeting in neuronal cell specification
Journal of Structural Biology: X :100043 (2020)
Smith N, Wilkinson-White L, Kwan A, Trewhella J, Matthews J
|
| RgGuinier |
3.3 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
39 |
nm3 |
|
|
|
|
|
|
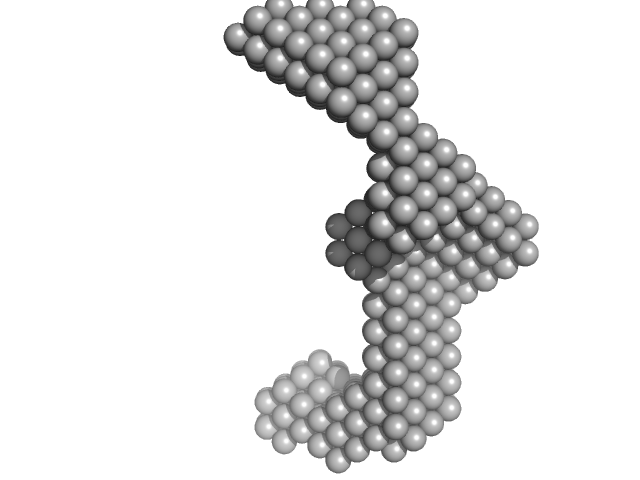
|
| Sample: |
Insulin gene enhancer protein ISL-1 monomer, 12 kDa Mus musculus protein
LIM/homeobox protein Lhx3 monomer, 9 kDa Mus musculus protein
|
| Buffer: |
20 mM sodium phosphate monobasic/dibasic, 100 mM NaCl, 1 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2018 Nov 2
|
Contrasting DNA-binding behaviour by ISL1 and LHX3 underpins differential gene targeting in neuronal cell specification
Journal of Structural Biology: X :100043 (2020)
Smith N, Wilkinson-White L, Kwan A, Trewhella J, Matthews J
|
| RgGuinier |
3.4 |
nm |
| Dmax |
12.7 |
nm |
| VolumePorod |
40 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Insulin gene enhancer protein ISL-1 monomer, 14 kDa Mus musculus protein
LIM/homeobox protein Lhx3 monomer, 23 kDa Mus musculus protein
|
| Buffer: |
20 mM sodium phosphate monobasic/dibasic, 100 mM NaCl, 1 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2018 Nov 2
|
Contrasting DNA-binding behaviour by ISL1 and LHX3 underpins differential gene targeting in neuronal cell specification
Journal of Structural Biology: X :100043 (2020)
Smith N, Wilkinson-White L, Kwan A, Trewhella J, Matthews J
|
| RgGuinier |
3.4 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
76 |
nm3 |
|
|