|
|
|
|
|
| Sample: |
Human Telomere Repeat (TTAGGG)12 monomer, 23 kDa synthetic construct DNA
|
| Buffer: |
6 mM Na2HPO4, 2 mM NaH2PO4, 1 mM Na2EDTA, 185 mM KCl, pH: 7.2 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2019 Aug 14
|
The solution structures of higher-order human telomere G-quadruplex multimers.
Nucleic Acids Res (2021)
Monsen RC, Chakravarthy S, Dean WL, Chaires JB, Trent JO
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.7 |
nm |
| VolumePorod |
26 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Human Telomere 96mer monomer, 31 kDa synthetic construct DNA
|
| Buffer: |
6 mM Na2HPO4, 2 mM NaH2PO4, 1 mM Na2EDTA, 185 mM KCl, pH: 7.2 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Feb 19
|
The solution structures of higher-order human telomere G-quadruplex multimers.
Nucleic Acids Res (2021)
Monsen RC, Chakravarthy S, Dean WL, Chaires JB, Trent JO
|
| RgGuinier |
3.2 |
nm |
| Dmax |
10.9 |
nm |
| VolumePorod |
38 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Di-domain acyl carrier protein of PigH from prodigiosin biosynthesis monomer, 22 kDa Serratia sp. ATCC … protein
|
| Buffer: |
20 mM Tris supplemented with 5 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at BL1.3W, Synchrotron Light Research Institute (SLRI) on 2016 Jan 13
|
Solution Structure and Conformational Dynamics of a Doublet Acyl Carrier Protein from Prodigiosin Biosynthesis.
Biochemistry (2021)
Thongkawphueak T, Winter AJ, Williams C, Maple HJ, Soontaranon S, Kaewhan C, Campopiano DJ, Crump MP, Wattana-Amorn P
|
| RgGuinier |
2.4 |
nm |
| Dmax |
8.4 |
nm |
| VolumePorod |
39 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Adagio protein 1 (Zeitlupe G46S:G80R) dimer, 37 kDa Arabidopsis thaliana protein
|
| Buffer: |
50 mM HEPES, 100 mM NaCl, 2 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2017 Nov 2
|
Steric and Electronic Interactions at Gln154 in ZEITLUPE Induce Reorganization of the LOV Domain Dimer Interface.
Biochemistry (2020)
Pudasaini A, Green R, Song YH, Blumenfeld A, Karki N, Imaizumi T, Zoltowski BD
|
| RgGuinier |
2.2 |
nm |
| Dmax |
8.4 |
nm |
| VolumePorod |
41 |
nm3 |
|
|
|
|
|
|

|
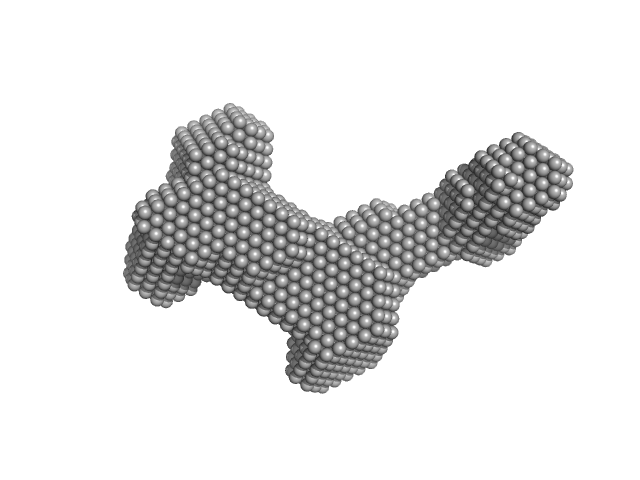
| Sample: |
Evolutionarily conserved signaling intermediate in Toll pathway, mitochondrial dimer, 44 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 250 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Dec 7
|
Assembly of the mitochondrial Complex I assembly complex suggests a regulatory role for deflavination.
Angew Chem Int Ed Engl (2020)
Giachin G, Jessop M, Bouverot R, Acajjaoui S, Saidi M, Chretien A, Bacia-Verloop M, Signor L, Mas PJ, Favier A, Borel Meneroud E, Hons M, Hart DJ, Kandiah E, Boeri Erba E, Buisson A, Leonard G, Gutsche I, Soler-Lopez M
|
| RgGuinier |
3.7 |
nm |
| Dmax |
11.9 |
nm |
| VolumePorod |
117 |
nm3 |
|
|
|
|
|
|
|
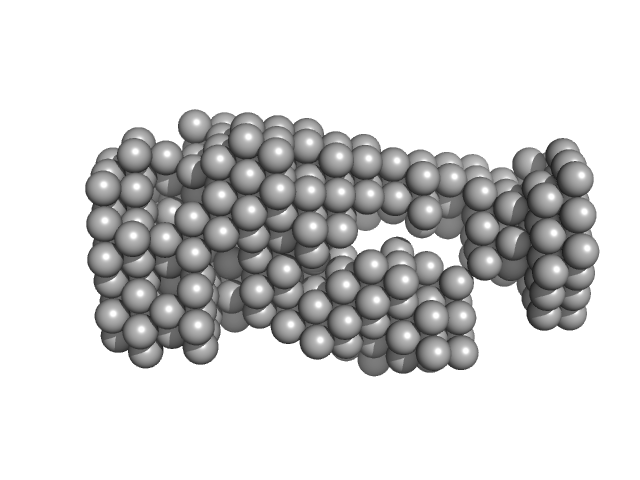
| Sample: |
Complex I intermediate-associated protein 30, mitochondrial monomer, 35 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 250 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 May 29
|
Assembly of the mitochondrial Complex I assembly complex suggests a regulatory role for deflavination.
Angew Chem Int Ed Engl (2020)
Giachin G, Jessop M, Bouverot R, Acajjaoui S, Saidi M, Chretien A, Bacia-Verloop M, Signor L, Mas PJ, Favier A, Borel Meneroud E, Hons M, Hart DJ, Kandiah E, Boeri Erba E, Buisson A, Leonard G, Gutsche I, Soler-Lopez M
|
| RgGuinier |
3.3 |
nm |
| Dmax |
9.6 |
nm |
| VolumePorod |
82 |
nm3 |
|
|
|
|
|
|
|
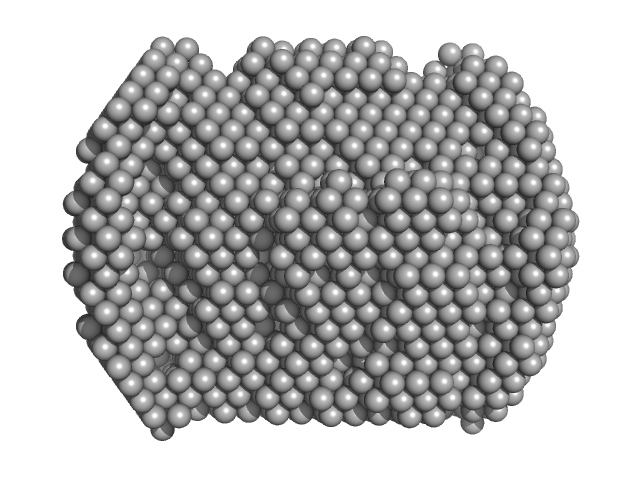
| Sample: |
Complex I assembly factor ACAD9, mitochondrial dimer, 132 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 250 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 May 28
|
Assembly of the mitochondrial Complex I assembly complex suggests a regulatory role for deflavination.
Angew Chem Int Ed Engl (2020)
Giachin G, Jessop M, Bouverot R, Acajjaoui S, Saidi M, Chretien A, Bacia-Verloop M, Signor L, Mas PJ, Favier A, Borel Meneroud E, Hons M, Hart DJ, Kandiah E, Boeri Erba E, Buisson A, Leonard G, Gutsche I, Soler-Lopez M
|
| RgGuinier |
3.6 |
nm |
| Dmax |
10.2 |
nm |
| VolumePorod |
217 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Complex I assembly factor ACAD9-VLCAD chimera dimer, 131 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 250 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Nov 30
|
Assembly of the mitochondrial Complex I assembly complex suggests a regulatory role for deflavination.
Angew Chem Int Ed Engl (2020)
Giachin G, Jessop M, Bouverot R, Acajjaoui S, Saidi M, Chretien A, Bacia-Verloop M, Signor L, Mas PJ, Favier A, Borel Meneroud E, Hons M, Hart DJ, Kandiah E, Boeri Erba E, Buisson A, Leonard G, Gutsche I, Soler-Lopez M
|
| RgGuinier |
3.9 |
nm |
| Dmax |
11.7 |
nm |
| VolumePorod |
217 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Very long-chain specific acyl-CoA dehydrogenase, mitochondrial dimer, 126 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 250 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Dec 2
|
Assembly of the mitochondrial Complex I assembly complex suggests a regulatory role for deflavination.
Angew Chem Int Ed Engl (2020)
Giachin G, Jessop M, Bouverot R, Acajjaoui S, Saidi M, Chretien A, Bacia-Verloop M, Signor L, Mas PJ, Favier A, Borel Meneroud E, Hons M, Hart DJ, Kandiah E, Boeri Erba E, Buisson A, Leonard G, Gutsche I, Soler-Lopez M
|
| RgGuinier |
3.4 |
nm |
| Dmax |
9.9 |
nm |
| VolumePorod |
215 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Complex I assembly factor ACAD9, mitochondrial dimer, 132 kDa Homo sapiens protein
Evolutionarily conserved signaling intermediate in Toll pathway, mitochondrial tetramer, 88 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 250 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Oct 4
|
Assembly of the mitochondrial Complex I assembly complex suggests a regulatory role for deflavination.
Angew Chem Int Ed Engl (2020)
Giachin G, Jessop M, Bouverot R, Acajjaoui S, Saidi M, Chretien A, Bacia-Verloop M, Signor L, Mas PJ, Favier A, Borel Meneroud E, Hons M, Hart DJ, Kandiah E, Boeri Erba E, Buisson A, Leonard G, Gutsche I, Soler-Lopez M
|
| RgGuinier |
6.0 |
nm |
| Dmax |
16.5 |
nm |
| VolumePorod |
683 |
nm3 |
|
|