|
|
|
|

|
| Sample: |
M100 oligonucleotide monomer, 12 kDa DNA
LIM/homeobox protein Lhx3 monomer, 10 kDa Mus musculus protein
|
| Buffer: |
20 mM sodium phosphate monobasic/dibasic, 100 mM NaCl, 1 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2018 Nov 2
|
Contrasting DNA-binding behaviour by ISL1 and LHX3 underpins differential gene targeting in neuronal cell specification
Journal of Structural Biology: X :100043 (2020)
Smith N, Wilkinson-White L, Kwan A, Trewhella J, Matthews J
|
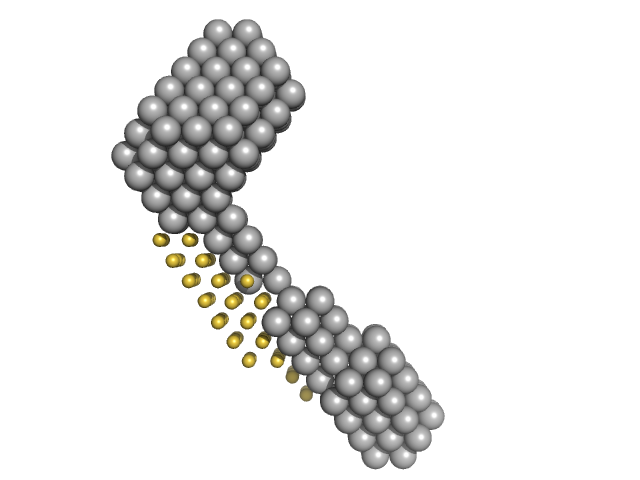
| RgGuinier |
2.1 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
27 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
M100 oligonucleotide monomer, 12 kDa DNA
LIM/homeobox protein Lhx3 monomer, 23 kDa Mus musculus protein
Insulin gene enhancer protein ISL-1 monomer, 4 kDa Mus musculus protein
|
| Buffer: |
20 mM sodium phosphate monobasic/dibasic, 100 mM NaCl, 1 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2018 Nov 2
|
Contrasting DNA-binding behaviour by ISL1 and LHX3 underpins differential gene targeting in neuronal cell specification
Journal of Structural Biology: X :100043 (2020)
Smith N, Wilkinson-White L, Kwan A, Trewhella J, Matthews J
|
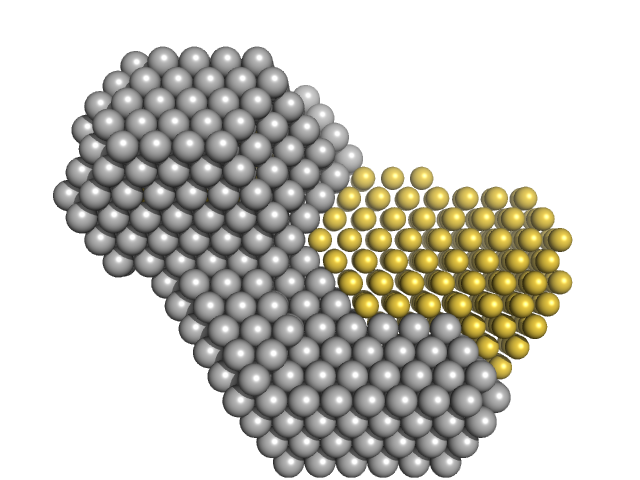
| RgGuinier |
3.6 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
49 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
M100 oligonucleotide monomer, 12 kDa DNA
Insulin gene enhancer protein ISL-1 monomer, 12 kDa Mus musculus protein
LIM/homeobox protein Lhx3 monomer, 9 kDa Mus musculus protein
|
| Buffer: |
20 mM sodium phosphate monobasic/dibasic, 100 mM NaCl, 1 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2018 Nov 2
|
Contrasting DNA-binding behaviour by ISL1 and LHX3 underpins differential gene targeting in neuronal cell specification
Journal of Structural Biology: X :100043 (2020)
Smith N, Wilkinson-White L, Kwan A, Trewhella J, Matthews J
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
41 |
nm3 |
|
|
|
|
|
|
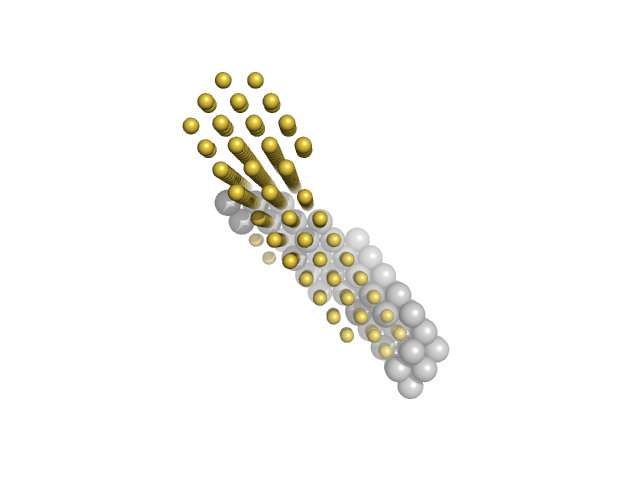
|
| Sample: |
M100 oligonucleotide monomer, 12 kDa DNA
Insulin gene enhancer protein ISL-1 monomer, 14 kDa Mus musculus protein
LIM/homeobox protein Lhx3 monomer, 23 kDa Mus musculus protein
|
| Buffer: |
20 mM sodium phosphate monobasic/dibasic, 100 mM NaCl, 1 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2018 Nov 2
|
Contrasting DNA-binding behaviour by ISL1 and LHX3 underpins differential gene targeting in neuronal cell specification
Journal of Structural Biology: X :100043 (2020)
Smith N, Wilkinson-White L, Kwan A, Trewhella J, Matthews J
|
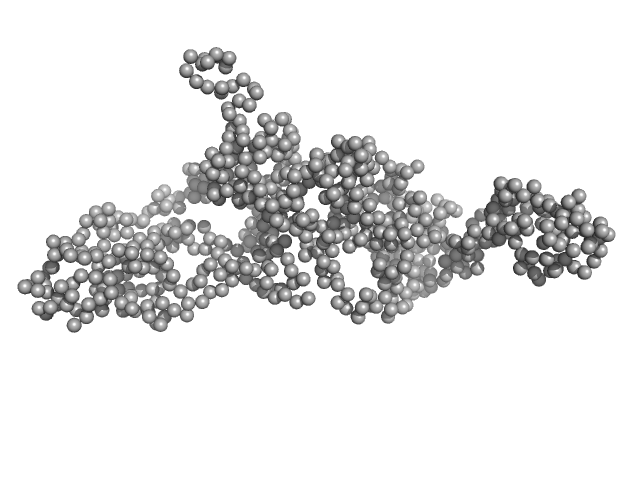
| RgGuinier |
3.6 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
70 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
M100 oligonucleotide monomer, 12 kDa DNA
|
| Buffer: |
20 mM sodium phosphate monobasic/dibasic, 100 mM NaCl, 1 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2018 Nov 2
|
Contrasting DNA-binding behaviour by ISL1 and LHX3 underpins differential gene targeting in neuronal cell specification
Journal of Structural Biology: X :100043 (2020)
Smith N, Wilkinson-White L, Kwan A, Trewhella J, Matthews J
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.8 |
nm |
| VolumePorod |
15 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Modular nanotransporter with an epidermal growth factor ligand module monomer, 76 kDa synthetic construct protein
|
| Buffer: |
150 mM NaCl, 10 mM phosphate buffer saline, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Nov 24
|
Low-resolution structures of modular nanotransporters shed light on their functional activity
Acta Crystallographica Section D Structural Biology 76(12):1270-1279 (2020)
Khramtsov Y, Vlasova A, Vlasov A, Rosenkranz A, Ulasov A, Ryzhykau Y, Kuklin A, Orekhov A, Eydlin I, Georgiev G, Gordeliy V, Sobolev A
|
| RgGuinier |
4.2 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
161 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Primary microRNA pri-miR16-1 monomer, 36 kDa Homo sapiens RNA
|
| Buffer: |
50 mM KCl, 50 mM HEPES, 5 mM DTT, 1% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2017 Apr 12
|
Elucidating the Role of Microprocessor Protein DGCR8 in Bending RNA Structures
Biophysical Journal (2020)
Pabit S, Chen Y, Usher E, Cook E, Pollack L, Showalter S
|
| RgGuinier |
4.4 |
nm |
| Dmax |
17.2 |
nm |
| VolumePorod |
74 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Primary microRNA pri-miR16-1 complexed with DGCR8-core protein monomer, 36 kDa Homo sapiens RNA
Microprocessor complex subunit DGCR8 monomer, 26 kDa Homo sapiens protein
|
| Buffer: |
50 mM KCl, 50 mM HEPES, 5 mM DTT, 1% glycerol, 50% sucrose, DGCR8-core, pH: 7.5 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2017 Apr 12
|
Elucidating the Role of Microprocessor Protein DGCR8 in Bending RNA Structures
Biophysical Journal (2020)
Pabit S, Chen Y, Usher E, Cook E, Pollack L, Showalter S
|
| RgGuinier |
5.0 |
nm |
| Dmax |
17.2 |
nm |
| VolumePorod |
125 |
nm3 |
|
|
|
|
|
|
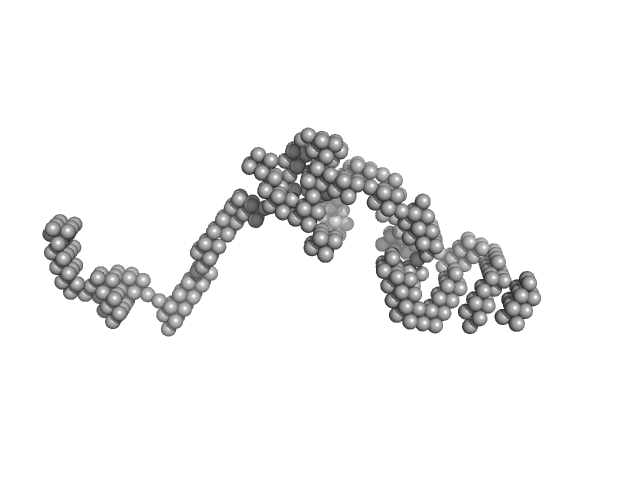
|
| Sample: |
Fibrillin-1 monomer, 44 kDa Homo sapiens protein
Elastin monomer, 60 kDa Homo sapiens protein
|
| Buffer: |
Tris buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Feb 21
|
Transglutaminase-Mediated Cross-Linking of Tropoelastin to Fibrillin Stabilises the Elastin Precursor Prior to Elastic Fibre Assembly.
J Mol Biol 432(21):5736-5751 (2020)
Lockhart-Cairns MP, Newandee H, Thomson J, Weiss AS, Baldock C, Tarakanova A
|
| RgGuinier |
7.1 |
nm |
| Dmax |
23.5 |
nm |
|
|
|
|
|
|

|
| Sample: |
HSP40, subfamily A dimer, 97 kDa Plasmodium falciparum protein
|
| Buffer: |
50 mM Tris, 5 mM MgCl2, 300 mM KCl, 1 mM β-mercaptoethanol, 5% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR-Central Drug Research Institute on 2019 Oct 24
|
Structural-functional diversity of malaria parasite's PfHSP70-1 and PfHSP40 chaperone pair gives an edge over human orthologs in chaperone-assisted protein folding.
Biochem J 477(18):3625-3643 (2020)
Anas M, Shukla A, Tripathi A, Kumari V, Prakash C, Nag P, Kumar LS, Sharma SK, Ramachandran R, Kumar N
|
| RgGuinier |
4.3 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
339 |
nm3 |
|
|