|
|
|
|

|
| Sample: |
Type 3 secretion system pilotin dimer, 19 kDa Salmonella enterica subsp. … protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-2000, University of British Columbia on 2017 Mar 23
|
Characterization of the Pilotin-Secretin Complex from the Salmonella enterica Type III Secretion System Using Hybrid Structural Methods.
Structure (2020)
Majewski DD, Okon M, Heinkel F, Robb CS, Vuckovic M, McIntosh LP, Strynadka NCJ
|
| RgGuinier |
2.1 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
29 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Vitamin K-dependent protein C monomer, 62 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 145 mM NaCl, 5 mM CaCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2017 Jul 14
|
Zymogen and activated protein C have similar structural architecture.
J Biol Chem (2020)
Stojanovski BM, Pelc LA, Zuo X, Di Cera E
|
| RgGuinier |
3.6 |
nm |
| Dmax |
14.0 |
nm |
|
|
|
|
|
|
|
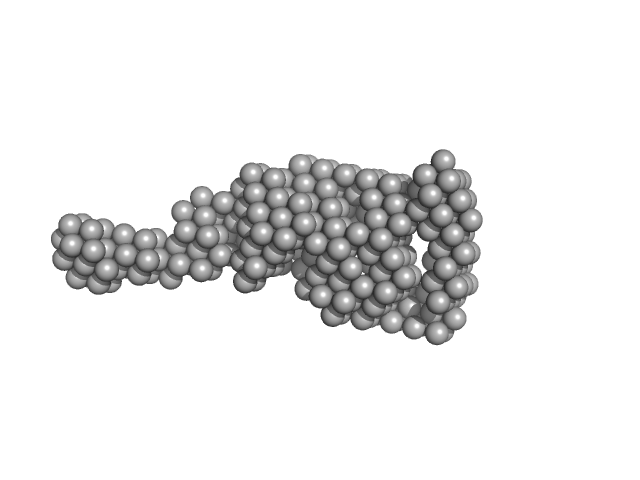
| Sample: |
Vitamin K-dependent protein C monomer, 62 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 145 mM NaCl, 5 mM CaCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2017 Aug 17
|
Zymogen and activated protein C have similar structural architecture.
J Biol Chem (2020)
Stojanovski BM, Pelc LA, Zuo X, Di Cera E
|
| RgGuinier |
3.7 |
nm |
| Dmax |
14.0 |
nm |
|
|
|
|
|
|

|
| Sample: |
Bifunctional hemolysin/adenylate cyclase monomer, 57 kDa Bordetella pertussis protein
|
| Buffer: |
10 mM Tris HCl, 150 mM NaCl, 10 mM CaCl₂, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Oct 31
|
Continuous Assembly of β-Roll Structures Is Implicated in the Type I-Dependent Secretion of Large Repeat-in-Toxins (RTX) Proteins.
J Mol Biol (2020)
Motlova L, Klimova N, Fiser R, Sebo P, Bumba L
|
| RgGuinier |
4.0 |
nm |
| Dmax |
13.3 |
nm |
| VolumePorod |
94 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Bifunctional hemolysin/adenylate cyclase monomer, 45 kDa Bordetella pertussis protein
|
| Buffer: |
10 mM Tris HCl, 150 mM NaCl, 10 mM CaCl₂, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Oct 31
|
Continuous Assembly of β-Roll Structures Is Implicated in the Type I-Dependent Secretion of Large Repeat-in-Toxins (RTX) Proteins.
J Mol Biol (2020)
Motlova L, Klimova N, Fiser R, Sebo P, Bumba L
|
| RgGuinier |
3.4 |
nm |
| Dmax |
12.1 |
nm |
| VolumePorod |
85 |
nm3 |
|
|
|
|
|
|

|
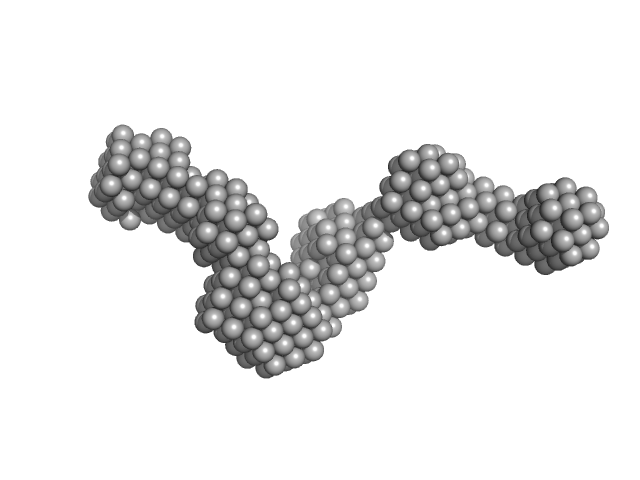
| Sample: |
Bifunctional hemolysin/adenylate cyclase monomer, 32 kDa Bordetella pertussis protein
|
| Buffer: |
10 mM Tris HCl, 150 mM NaCl, 10 mM CaCl₂, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Oct 31
|
Continuous Assembly of β-Roll Structures Is Implicated in the Type I-Dependent Secretion of Large Repeat-in-Toxins (RTX) Proteins.
J Mol Biol (2020)
Motlova L, Klimova N, Fiser R, Sebo P, Bumba L
|
| RgGuinier |
2.7 |
nm |
| Dmax |
8.6 |
nm |
| VolumePorod |
45 |
nm3 |
|
|
|
|
|
|
|
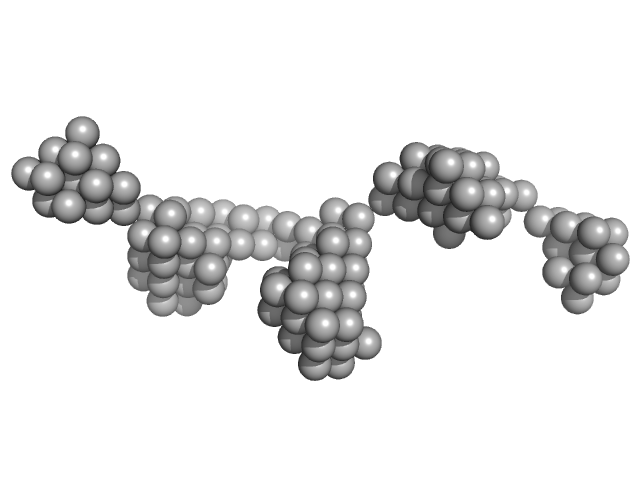
| Sample: |
Chitinase ChiA monomer, 70 kDa Flavobacterium johnsoniae protein
|
| Buffer: |
50 mM Tris, 250 mM NaCl, 0.25 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2019 May 7
|
Structural insights of the enzymes from the chitin utilization locus of Flavobacterium johnsoniae.
Sci Rep 10(1):13775 (2020)
Mazurkewich S, Helland R, Mackenzie A, Eijsink VGH, Pope PB, Brändén G, Larsbrink J
|
| RgGuinier |
4.7 |
nm |
| Dmax |
16.3 |
nm |
| VolumePorod |
99 |
nm3 |
|
|
|
|
|
|
|
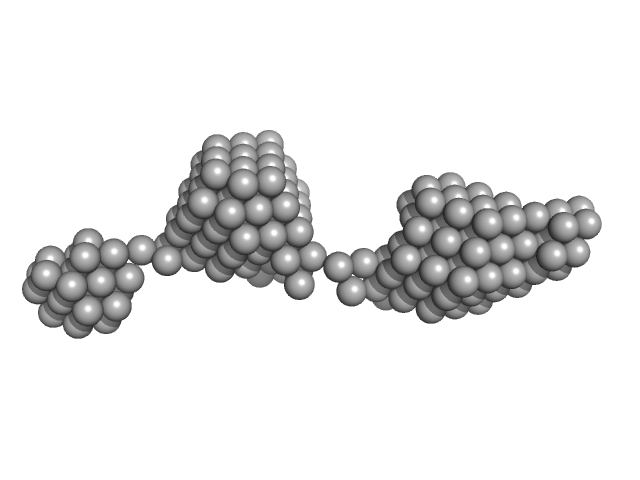
| Sample: |
Beta-ketoacyl synthase Bamb_5925 monomer, 4 kDa Burkholderia ambifaria protein
|
| Buffer: |
20 mM Tris-HCl, 200 mM NaCl, 5% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Jan 23
|
Towards improved understanding of intersubunit interactions in modular polyketide biosynthesis: docking in the enacyloxin IIa polyketide synthase.
J Struct Biol :107581 (2020)
Risser F, Collin S, Dos Santos-Morais R, Gruez A, Chagot B, Weissman KJ
|
| RgGuinier |
1.6 |
nm |
| Dmax |
6.2 |
nm |
| VolumePorod |
3 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Beta-ketoacyl synthase Bamb_5920 monomer, 2 kDa Burkholderia ambifaria protein
|
| Buffer: |
20 mM Tris-HCl, 200 mM NaCl, 5% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Jan 23
|
Towards improved understanding of intersubunit interactions in modular polyketide biosynthesis: docking in the enacyloxin IIa polyketide synthase.
J Struct Biol :107581 (2020)
Risser F, Collin S, Dos Santos-Morais R, Gruez A, Chagot B, Weissman KJ
|
| RgGuinier |
1.2 |
nm |
| Dmax |
4.9 |
nm |
| VolumePorod |
3 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Plasmodium falciparum Heat shock protein 90 dimer, 177 kDa Plasmodium falciparum protein
|
| Buffer: |
25 mM Tris-HCl, 100 mM KCl, 1 mM β-mercaptoethanol, 1 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS1 Beamline, Brazilian Synchrotron Light Laboratory on 2018 May 11
|
Solution structure of Plasmodium falciparum Hsp90 indicates a high flexible dimer
Archives of Biochemistry and Biophysics :108468 (2020)
Silva N, Torricillas M, Minari K, Barbosa L, Seraphim T, Borges J
|
| RgGuinier |
5.7 |
nm |
| Dmax |
19.0 |
nm |
| VolumePorod |
350 |
nm3 |
|
|