|
|
|
|

|
| Sample: |
Heat shock protein 70 monomer, 74 kDa Plasmodium falciparum protein
Peptide monomer, 1 kDa synthetic construct protein
|
| Buffer: |
50mM Tris, 5mM MgCl2, 500mM KCl, 5mM Bme, 5% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR-Central Drug Research Institute on 2019 Oct 24
|
Structural-functional diversity of malaria parasite's PfHSP70-1 and PfHSP40 chaperone pair gives an edge over human orthologs in chaperone-assisted protein folding.
Biochem J 477(18):3625-3643 (2020)
Anas M, Shukla A, Tripathi A, Kumari V, Prakash C, Nag P, Kumar LS, Sharma SK, Ramachandran R, Kumar N
|
| RgGuinier |
5.3 |
nm |
| Dmax |
16.6 |
nm |
| VolumePorod |
450 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Mce-family protein Mce4A monomer, 44 kDa Mycobacterium tuberculosis (strain … protein
N-Dodecyl-β-D-Maltopyranoside 0, 102 kDa
|
| Buffer: |
50mM Tris, 500mM NaCl, 10% Glycerol, 5mM DDM, 1mM Beta-ME, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 May 13
|
Structural insights into the substrate-binding proteins Mce1A and Mce4A from Mycobacterium tuberculosis
IUCrJ 8(5) (2021)
Asthana P, Singh D, Pedersen J, Hynönen M, Sulu R, Murthy A, Laitaoja M, Jänis J, Riley L, Venkatesan R
|
| RgGuinier |
5.7 |
nm |
| Dmax |
21.5 |
nm |
| VolumePorod |
446 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
N-Dodecyl-β-D-Maltopyranoside 0, 102 kDa
Mce-family protein Mce4A monomer, 35 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
50mM Tris, 500mM NaCl, 10% Glycerol, 5mM DDM, 1mM Beta-ME, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Nov 28
|
Structural insights into the substrate-binding proteins Mce1A and Mce4A from Mycobacterium tuberculosis
IUCrJ 8(5) (2021)
Asthana P, Singh D, Pedersen J, Hynönen M, Sulu R, Murthy A, Laitaoja M, Jänis J, Riley L, Venkatesan R
|
| RgGuinier |
5.0 |
nm |
| Dmax |
19.1 |
nm |
| VolumePorod |
278 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
N-Dodecyl-β-D-Maltopyranoside 0, 102 kDa
Mce-family protein Mce4A monomer, 34 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
50mM Tris, 500mM NaCl, 10% Glycerol, 5mM DDM, 1mM Beta-ME, pH: 8.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 May 2
|
Structural insights into the substrate-binding proteins Mce1A and Mce4A from Mycobacterium tuberculosis
IUCrJ 8(5) (2021)
Asthana P, Singh D, Pedersen J, Hynönen M, Sulu R, Murthy A, Laitaoja M, Jänis J, Riley L, Venkatesan R
|
| RgGuinier |
4.8 |
nm |
| Dmax |
18.2 |
nm |
| VolumePorod |
189 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Mce-family protein Mce1A monomer, 36 kDa Mycobacterium tuberculosis protein
N-Dodecyl-β-D-Maltopyranoside 0, 82 kDa
|
| Buffer: |
50 mM Tris, 500 mM NaCl, 10% Glycerol, 5 mM DDM, 1 mM β-ME, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 May 2
|
Structural insights into the substrate-binding proteins Mce1A and Mce4A from Mycobacterium tuberculosis
IUCrJ 8(5) (2021)
Asthana P, Singh D, Pedersen J, Hynönen M, Sulu R, Murthy A, Laitaoja M, Jänis J, Riley L, Venkatesan R
|
| RgGuinier |
4.6 |
nm |
| Dmax |
17.5 |
nm |
| VolumePorod |
151 |
nm3 |
|
|
|
|
|
|
|
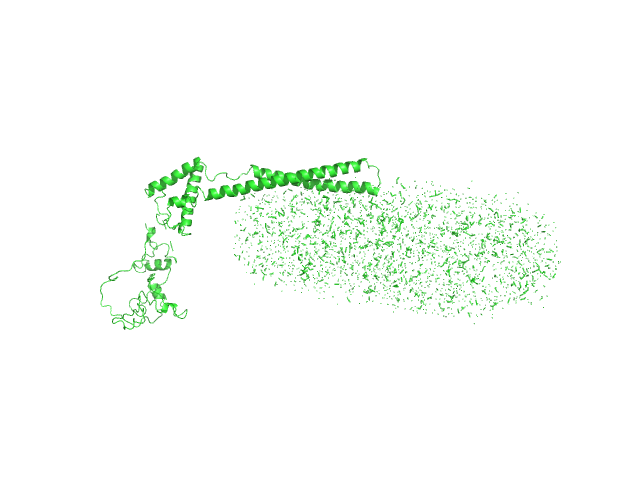
| Sample: |
Mce-family protein Mce1A monomer, 39 kDa Mycobacterium tuberculosis protein
N-Dodecyl-β-D-Maltopyranoside 0, 123 kDa
|
| Buffer: |
50 mM Tris, 350 mM NaCl, 10% Glycerol, 5mM DDM, 1 mM β-ME, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Nov 28
|
Structural insights into the substrate-binding proteins Mce1A and Mce4A from Mycobacterium tuberculosis
IUCrJ 8(5) (2021)
Asthana P, Singh D, Pedersen J, Hynönen M, Sulu R, Murthy A, Laitaoja M, Jänis J, Riley L, Venkatesan R
|
| RgGuinier |
5.5 |
nm |
| Dmax |
22.0 |
nm |
| VolumePorod |
216 |
nm3 |
|
|
|
|
|
|
|
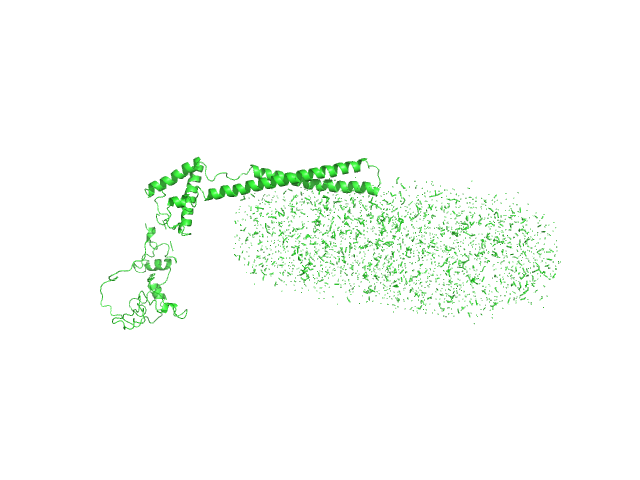
| Sample: |
Mce-family protein Mce1A monomer, 48 kDa Mycobacterium tuberculosis protein
N-Dodecyl-β-D-Maltopyranoside 0, 112 kDa
|
| Buffer: |
50 mM Tris, 500 mM NaCl, 10% Glycerol, 5mM DDM, 1 mM β-ME, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 May 13
|
Structural insights into the substrate-binding proteins Mce1A and Mce4A from Mycobacterium tuberculosis
IUCrJ 8(5) (2021)
Asthana P, Singh D, Pedersen J, Hynönen M, Sulu R, Murthy A, Laitaoja M, Jänis J, Riley L, Venkatesan R
|
| RgGuinier |
5.3 |
nm |
| Dmax |
21.6 |
nm |
| VolumePorod |
362 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Aldehyde-alcohol dehydrogenase dimer, 97 kDa Escherichia coli O157:H7 protein
|
| Buffer: |
20 mM Tris 400 mM NaCl 5% v/v glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2015 Nov 27
|
High-resolution structure of the alcohol dehydrogenase domain of the bifunctional bacterial enzyme AdhE.
Acta Crystallogr F Struct Biol Commun 76(Pt 9):414-421 (2020)
Azmi L, Bragginton EC, Cadby IT, Byron O, Roe AJ, Lovering AL, Gabrielsen M
|
| RgGuinier |
3.2 |
nm |
| Dmax |
16.9 |
nm |
| VolumePorod |
146 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Aldehyde-alcohol dehydrogenase monomer, 48 kDa Escherichia coli O157:H7 protein
|
| Buffer: |
30 mM HEPES, 150 mM NaCl, 5% (v/v) glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Jan 30
|
High-resolution structure of the alcohol dehydrogenase domain of the bifunctional bacterial enzyme AdhE.
Acta Crystallogr F Struct Biol Commun 76(Pt 9):414-421 (2020)
Azmi L, Bragginton EC, Cadby IT, Byron O, Roe AJ, Lovering AL, Gabrielsen M
|
| RgGuinier |
2.7 |
nm |
| Dmax |
11.1 |
nm |
| VolumePorod |
74 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Type 3 secretion system pilotin dimer, 28 kDa Salmonella enterica subsp. … protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-2000, University of British Columbia on 2017 Mar 23
|
Characterization of the Pilotin-Secretin Complex from the Salmonella enterica Type III Secretion System Using Hybrid Structural Methods.
Structure (2020)
Majewski DD, Okon M, Heinkel F, Robb CS, Vuckovic M, McIntosh LP, Strynadka NCJ
|
| RgGuinier |
3.1 |
nm |
| Dmax |
12.6 |
nm |
| VolumePorod |
53 |
nm3 |
|
|