|
|
|
|
|
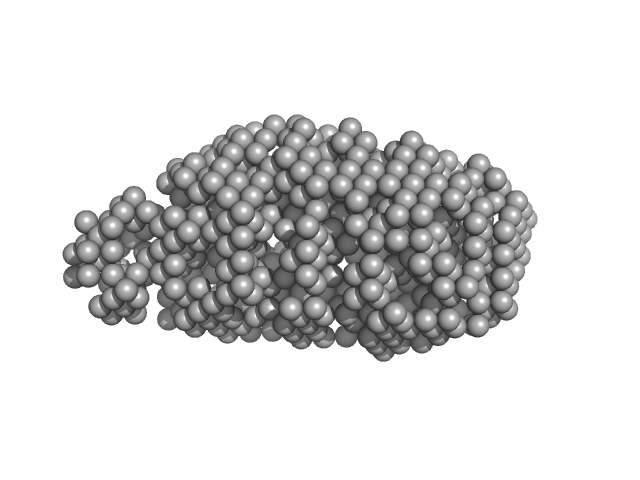
| Sample: |
Serine acetyltransferase hexamer, 177 kDa Escherichia coli protein
|
| Buffer: |
20 mM sodium phosphate, 85 mM NaCl, 2 mM EDTA, 10 mM 2-MCE, pH: 7.5 |
| Experiment: |
SAXS
data collected at Austrian SAXS beamline 5.2L, ELETTRA on 2016 Jun 1
|
Combination of SAXS and Protein Painting Discloses the Three-Dimensional Organization of the Bacterial Cysteine Synthase Complex, a Potential Target for Enhancers of Antibiotic Action.
Int J Mol Sci 20(20) (2019)
Rosa B, Marchetti M, Paredi G, Amenitsch H, Franko N, Benoni R, Giabbai B, De Marino MG, Mozzarelli A, Ronda L, Storici P, Campanini B, Bettati S
|
| RgGuinier |
3.9 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
280 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Cysteine synthase A (4-mer) tetramer, 143 kDa Escherichia coli protein
Serine acetyltransferase (6-mer) hexamer, 177 kDa Escherichia coli protein
|
| Buffer: |
20 mM sodium phosphate, 85 mM NaCl, 2 mM EDTA, 10 mM 2-MCE, pH: 7.5 |
| Experiment: |
SAXS
data collected at Austrian SAXS beamline 5.2L, ELETTRA on 2016 Jun 1
|
Combination of SAXS and Protein Painting Discloses the Three-Dimensional Organization of the Bacterial Cysteine Synthase Complex, a Potential Target for Enhancers of Antibiotic Action.
Int J Mol Sci 20(20) (2019)
Rosa B, Marchetti M, Paredi G, Amenitsch H, Franko N, Benoni R, Giabbai B, De Marino MG, Mozzarelli A, Ronda L, Storici P, Campanini B, Bettati S
|
| RgGuinier |
6.1 |
nm |
| Dmax |
22.0 |
nm |
| VolumePorod |
457 |
nm3 |
|
|
|
|
|
|
|
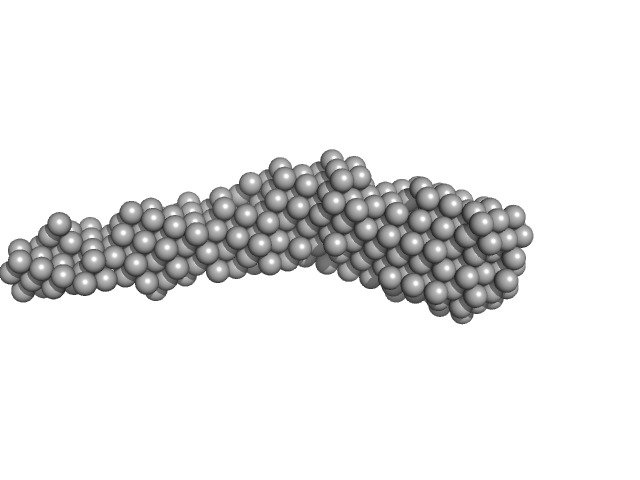
| Sample: |
Flagella binding tail protein monomer, 103 kDa Salmonella virus Chi protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 0.03 % NaN3, 5.0 % glycerol, pH: 7.8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Apr 4
|
The flagellotropic bacteriophage YSD1 targets Salmonella Typhi with a Chi-like protein tail fibre.
Mol Microbiol (2019)
Dunstan RA, Pickard D, Dougan S, Goulding D, Cormie C, Hardy J, Li F, Grinter R, Harcourt K, Yu L, Song J, Schreiber F, Choudhary J, Clare S, Coulibaly F, Strugnell RA, Dougan G, Lithgow T
|
| RgGuinier |
5.6 |
nm |
| Dmax |
27.4 |
nm |
| VolumePorod |
155 |
nm3 |
|
|
|
|
|
|

|
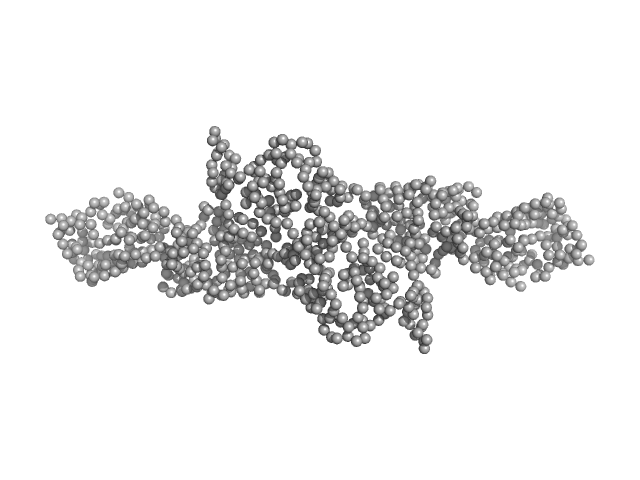
| Sample: |
Cytohesin-2; ARNO truncation mutant monomer, 40 kDa Homo sapiens protein
|
| Buffer: |
300 mM NaCl, 2 mM 2-mercaptoethanol and 30 mM Tris-HCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2015 Nov 25
|
Structural Organization and Dynamics of Homodimeric Cytohesin Family Arf GTPase Exchange Factors in Solution and on Membranes.
Structure (2019)
Das S, Malaby AW, Nawrotek A, Zhang W, Zeghouf M, Maslen S, Skehel M, Chakravarthy S, Irving TC, Bilsel O, Cherfils J, Lambright DG
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.9 |
nm |
| VolumePorod |
63 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Cytohesin-2 ARF nucleotide-binding site opener dimer, 93 kDa Homo sapiens protein
|
| Buffer: |
300 mM NaCl, 2 mM 2-mercaptoethanol and 30 mM Tris-HCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Jun 23
|
Structural Organization and Dynamics of Homodimeric Cytohesin Family Arf GTPase Exchange Factors in Solution and on Membranes.
Structure (2019)
Das S, Malaby AW, Nawrotek A, Zhang W, Zeghouf M, Maslen S, Skehel M, Chakravarthy S, Irving TC, Bilsel O, Cherfils J, Lambright DG
|
| RgGuinier |
4.8 |
nm |
| Dmax |
19.7 |
nm |
| VolumePorod |
145 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Cytohesin-3 dimer, 93 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 2 mM MgCl2, 0.1% 2-mercaptoethanol, 5% glycerol, 0.001 mM insitol 1,3,4,5-tetrakis phosphate, pH: 8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2013 Nov 15
|
Structural Organization and Dynamics of Homodimeric Cytohesin Family Arf GTPase Exchange Factors in Solution and on Membranes.
Structure (2019)
Das S, Malaby AW, Nawrotek A, Zhang W, Zeghouf M, Maslen S, Skehel M, Chakravarthy S, Irving TC, Bilsel O, Cherfils J, Lambright DG
|
| RgGuinier |
5.5 |
nm |
| Dmax |
26.0 |
nm |
| VolumePorod |
194 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Cytohesin-3 dimer, 90 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 2 mM MgCl2, 0.1% 2-mercaptoethanol, 5% glycerol, 0.001 mM insitol 1,3,4,5-tetrakis phosphate, pH: 8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2013 Nov 15
|
Structural Organization and Dynamics of Homodimeric Cytohesin Family Arf GTPase Exchange Factors in Solution and on Membranes.
Structure (2019)
Das S, Malaby AW, Nawrotek A, Zhang W, Zeghouf M, Maslen S, Skehel M, Chakravarthy S, Irving TC, Bilsel O, Cherfils J, Lambright DG
|
| RgGuinier |
5.3 |
nm |
| Dmax |
25.7 |
nm |
| VolumePorod |
168 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
4-hydroxy-tetrahydrodipicolinate synthase tetramer, 131 kDa Campylobacter jejuni protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Aug 2
|
Asparagine-84, a regulatory allosteric site residue, helps maintain the quaternary structure of Campylobacter jejuni dihydrodipicolinate synthase.
J Struct Biol :107409 (2019)
Majdi Yazdi M, Saran S, Mrozowich T, Lehnert C, Patel TR, Sanders DAR, Palmer DRJ
|
| RgGuinier |
3.4 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
188 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
4-hydroxy-tetrahydrodipicolinate synthase (N84D mutant) dimer, 65 kDa Campylobacter jejuni protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Aug 2
|
Asparagine-84, a regulatory allosteric site residue, helps maintain the quaternary structure of Campylobacter jejuni dihydrodipicolinate synthase.
J Struct Biol :107409 (2019)
Majdi Yazdi M, Saran S, Mrozowich T, Lehnert C, Patel TR, Sanders DAR, Palmer DRJ
|
| RgGuinier |
3.1 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
108 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
4-hydroxy-tetrahydrodipicolinate synthase (N84A mutant), 33 kDa Campylobacter jejuni protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Aug 2
|
Asparagine-84, a regulatory allosteric site residue, helps maintain the quaternary structure of Campylobacter jejuni dihydrodipicolinate synthase.
J Struct Biol :107409 (2019)
Majdi Yazdi M, Saran S, Mrozowich T, Lehnert C, Patel TR, Sanders DAR, Palmer DRJ
|
| RgGuinier |
3.3 |
nm |
| Dmax |
8.9 |
nm |
| VolumePorod |
163 |
nm3 |
|
|