|
|
|
|
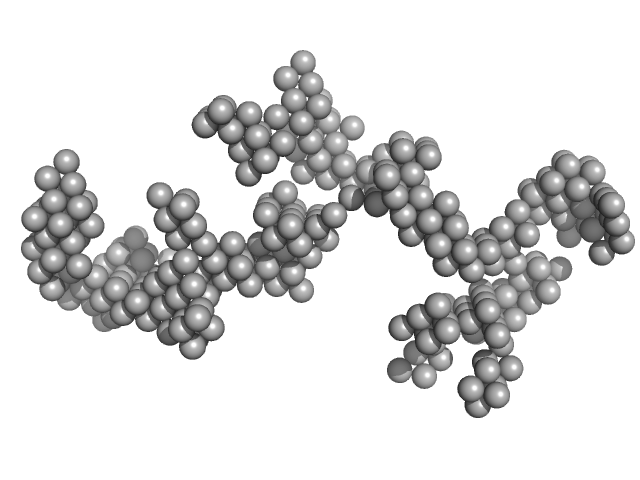
|
| Sample: |
Braveheart RNA monomer, 205 kDa Homo sapiens RNA
Cellular nucleic acid-binding protein monomer, 22 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES-KOH, 100 mM KCl, 6 mM MgCl2, pH: 7.6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Jun 23
|
Zinc-finger protein CNBP alters the 3-D structure of lncRNA Braveheart in solution
Nature Communications 11(1) (2020)
Kim D, Thiel B, Mrozowich T, Hennelly S, Hofacker I, Patel T, Sanbonmatsu K
|
| RgGuinier |
9.8 |
nm |
| Dmax |
30.2 |
nm |
| VolumePorod |
1660 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Cellular nucleic acid-binding protein monomer, 22 kDa Homo sapiens protein
Braveheart Fragment 1 monomer, 116 kDa Homo sapiens RNA
|
| Buffer: |
50 mM HEPES-KOH, 100 mM KCl, 6 mM MgCl2, pH: 7.6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Jun 23
|
Zinc-finger protein CNBP alters the 3-D structure of lncRNA Braveheart in solution
Nature Communications 11(1) (2020)
Kim D, Thiel B, Mrozowich T, Hennelly S, Hofacker I, Patel T, Sanbonmatsu K
|
| RgGuinier |
8.2 |
nm |
| Dmax |
27.0 |
nm |
| VolumePorod |
455 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Resistance to inhibitors of cholinesterase 8 homolog A monomer, 56 kDa Rattus norvegicus protein
Guanine nucleotide-binding protein G(i) subunit alpha-1 monomer, 38 kDa Rattus norvegicus protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2019 Jul 30
|
Structure of the G protein chaperone and guanine nucleotide exchange factor Ric-8A bound to Gαi1
Nature Communications 11(1) (2020)
McClelland L, Zhang K, Mou T, Johnston J, Yates-Hansen C, Li S, Thomas C, Doukov T, Triest S, Wohlkonig A, Tall G, Steyaert J, Chiu W, Sprang S
|
| RgGuinier |
3.5 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
120 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
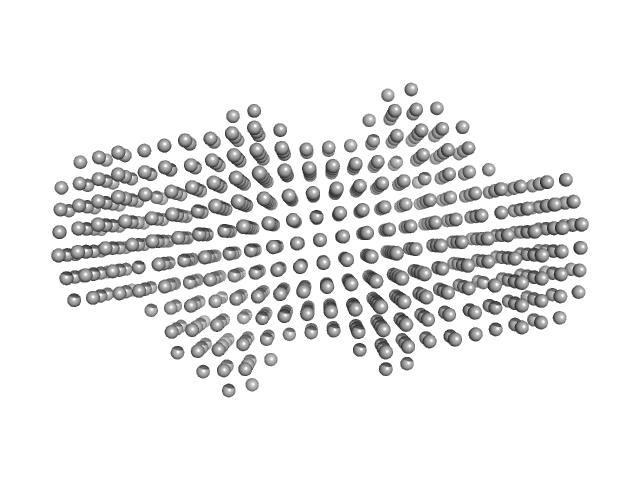
P. maculata perivitellin 2 dimer, 188 kDa Pomacea maculata protein
|
| Buffer: |
20 mM Tris, pH: 7 |
| Experiment: |
SAXS
data collected at SAXS2 Beamline, Brazilian Synchrotron Light Laboratory on 2015 Mar 26
|
Exaptation of two ancient immune proteins into a new dimeric pore-forming toxin in snails.
J Struct Biol 211(2):107531 (2020)
Giglio ML, Ituarte S, Milesi V, Dreon MS, Brola TR, Caramelo J, Ip JCH, Maté S, Qiu JW, Otero LH, Heras H
|
| RgGuinier |
4.4 |
nm |
| Dmax |
14.3 |
nm |
| VolumePorod |
267 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
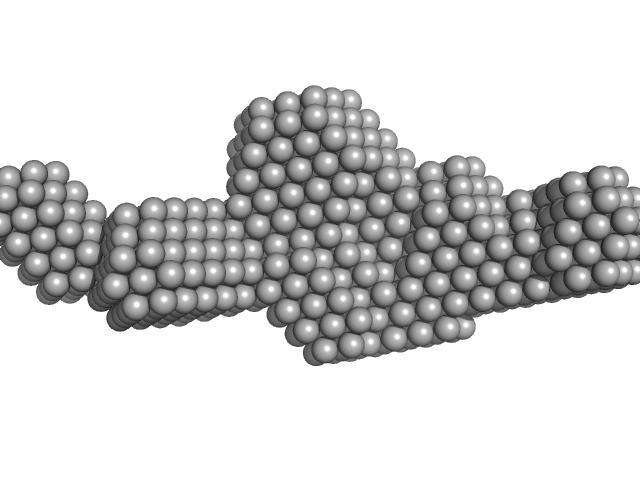
Sa0446 binding sequence 40bp monomer, 25 kDa DNA
Transcriptional regulator Lrs14-like protein dimer, 33 kDa Sulfolobus acidocaldarius protein
|
| Buffer: |
300 mM NaCl, 20 mM HEPES, pH 7.5, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Nov 5
|
Solution Structure of Archaeal Biofilm Regulator 2 (AbfR2) in Complex with 40 bp DNA
Marian Vogt
|
| RgGuinier |
3.4 |
nm |
| Dmax |
12.8 |
nm |
| VolumePorod |
60 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Endoribonuclease E tetramer, 247 kDa Escherichia coli protein
|
| Buffer: |
10 mM DTT, 10 mM MgCl2, 0.5 M NaCl, 20 mM Tris, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Feb 11
|
A structural and biochemical comparison of Ribonuclease E homologues from pathogenic bacteria highlights species-specific properties.
Sci Rep 9(1):7952 (2019)
Mardle CE, Shakespeare TJ, Butt LE, Goddard LR, Gowers DM, Atkins HS, Vincent HA, Callaghan AJ
|
| RgGuinier |
5.0 |
nm |
| Dmax |
16.1 |
nm |
| VolumePorod |
468 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
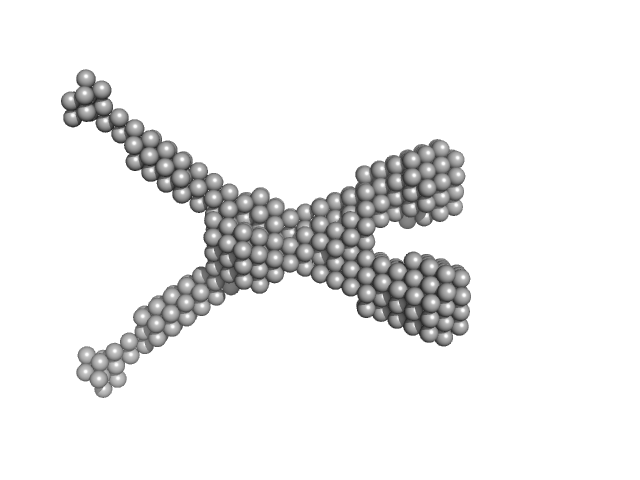
Noelin tetramer, 72 kDa Mus musculus protein
|
| Buffer: |
150 mM NaCl, 20 mM HEPES, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Feb 5
|
Design and structural characterisation of olfactomedin-1 variants as tools for functional studies.
BMC Mol Cell Biol 20(1):50 (2019)
Pronker MF, van den Hoek H, Janssen BJC
|
| RgGuinier |
5.4 |
nm |
| Dmax |
16.3 |
nm |
| VolumePorod |
160 |
nm3 |
|
|
|
|
|
|
|
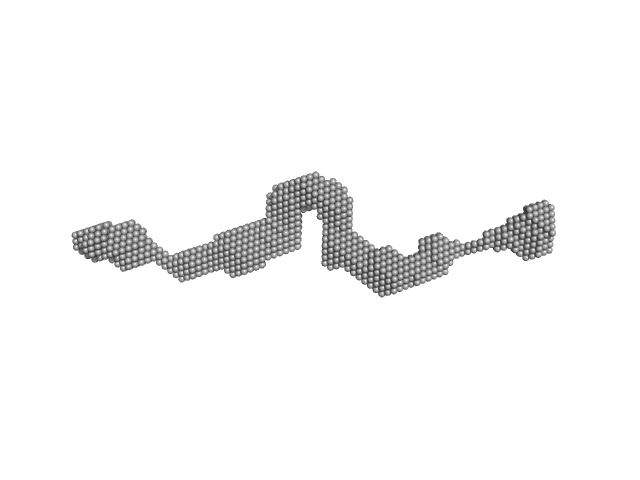
| Sample: |
Dystrophin (R11-15 human dystrophin fragment) monomer, 60 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-d11, 150 mM NaCl, 0.1 mM EDTA-d16, in 100% v/v D2O, pH: 7.1 |
| Experiment: |
SANS
data collected at D22, Institut Laue-Langevin (ILL) on 2016 Nov 1
|
How the central domain of dystrophin acts to bridge F-actin to sarcolemmal lipids.
J Struct Biol :107411 (2019)
Mias-Lucquin D, Dos Santos Morais R, Chéron A, Lagarrigue M, Winder SJ, Chenuel T, Pérez J, Appavou MS, Martel A, Alviset G, Le Rumeur E, Combet S, Hubert JF, Delalande O
|
| RgGuinier |
6.2 |
nm |
| Dmax |
28.1 |
nm |
| VolumePorod |
144 |
nm3 |
|
|
|
|
|
|

|
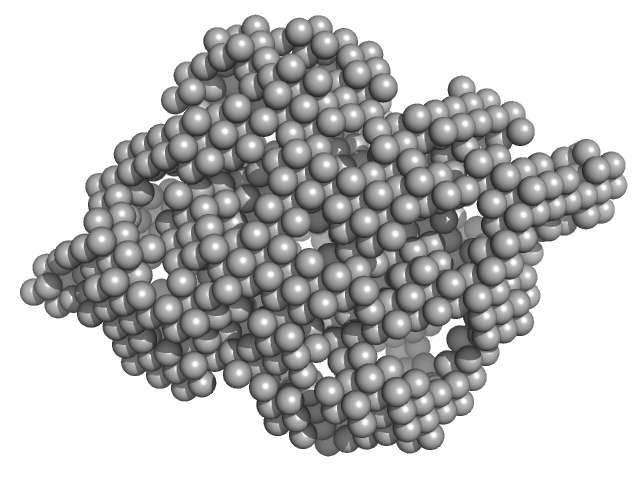
| Sample: |
Dystrophin (R11-15 human dystrophin fragment) monomer, 60 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-d11, 150 mM NaCl, 0.1 mM EDTA-d16, in 100% v/v D2O, pH: 7.1 |
| Experiment: |
SANS
data collected at D22, Institut Laue-Langevin (ILL) on 2016 Nov 1
|
How the central domain of dystrophin acts to bridge F-actin to sarcolemmal lipids.
J Struct Biol :107411 (2019)
Mias-Lucquin D, Dos Santos Morais R, Chéron A, Lagarrigue M, Winder SJ, Chenuel T, Pérez J, Appavou MS, Martel A, Alviset G, Le Rumeur E, Combet S, Hubert JF, Delalande O
|
| RgGuinier |
8.1 |
nm |
| Dmax |
29.6 |
nm |
| VolumePorod |
231 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Cysteine synthase A dimer, 71 kDa Escherichia coli protein
|
| Buffer: |
20 mM sodium phosphate, 85 mM NaCl, 2 mM EDTA, 10 mM 2-MCE, pH: 7.5 |
| Experiment: |
SAXS
data collected at Austrian SAXS beamline 5.2L, ELETTRA on 2016 Jun 1
|
Combination of SAXS and Protein Painting Discloses the Three-Dimensional Organization of the Bacterial Cysteine Synthase Complex, a Potential Target for Enhancers of Antibiotic Action.
Int J Mol Sci 20(20) (2019)
Rosa B, Marchetti M, Paredi G, Amenitsch H, Franko N, Benoni R, Giabbai B, De Marino MG, Mozzarelli A, Ronda L, Storici P, Campanini B, Bettati S
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
108 |
nm3 |
|
|