|
|
|
|
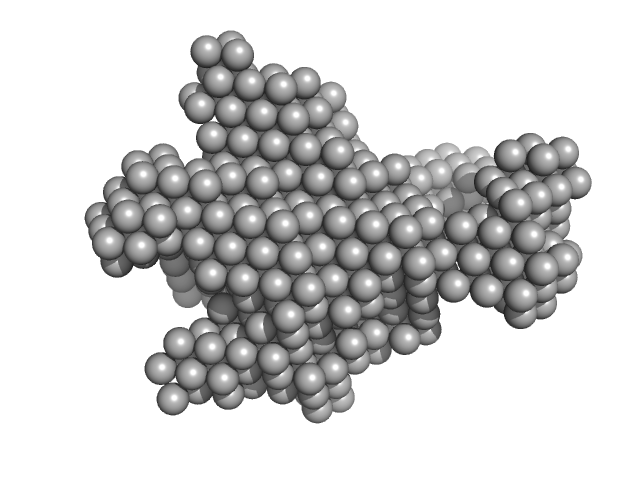
|
| Sample: |
Methylxanthine N1-demethylase NdmA trimer, 207 kDa Pseudomonas putida protein
Methylxanthine N3-demethylase NdmB trimer, 208 kDa Pseudomonas putida protein
|
| Buffer: |
20 mM HEPES 150 mM NaCl 2 mM TCEP 10% v/v glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at 4C, Pohang Accelerator Laboratory on 2018 Jul 27
|
Structural and Mechanistic Insights into Caffeine Degradation by the Bacterial N-Demethylase Complex.
J Mol Biol 431(19):3647-3661 (2019)
Kim JH, Kim BH, Brooks S, Kang SY, Summers RM, Song HK
|
| RgGuinier |
5.6 |
nm |
| Dmax |
19.1 |
nm |
|
|
|
|
|
|
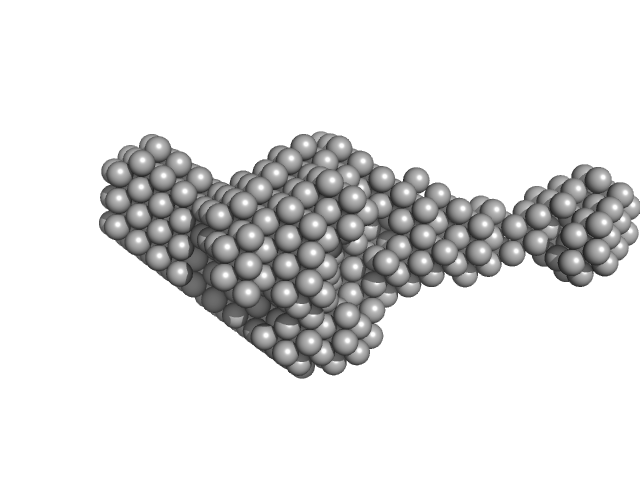
|
| Sample: |
Chitinase 2 monomer, 32 kDa Agave tequilana protein
|
| Buffer: |
MES 50 mM, pH: 6 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2017 Apr 19
|
A biophysical and structural study of two chitinases from Agave tequilana and their potential role as defense proteins.
FEBS J (2019)
Sierra-Gómez Y, Rodríguez-Hernández A, Cano-Sánchez P, Gómez-Velasco H, Hernández-Santoyo A, Siliqi D, Rodríguez-Romero A
|
| RgGuinier |
2.4 |
nm |
| Dmax |
9.8 |
nm |
| VolumePorod |
49 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Chitinase 1 monomer, 32 kDa Agave tequilana protein
|
| Buffer: |
MES 50 mM, pH: 6 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2017 Apr 26
|
A biophysical and structural study of two chitinases from Agave tequilana and their potential role as defense proteins.
FEBS J (2019)
Sierra-Gómez Y, Rodríguez-Hernández A, Cano-Sánchez P, Gómez-Velasco H, Hernández-Santoyo A, Siliqi D, Rodríguez-Romero A
|
| RgGuinier |
2.6 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
52 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
TraI monomer, 91 kDa Neisseria gonorrhoeae protein
|
| Buffer: |
50 mM TRIS-HCl 100 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Mar 5
|
DNA processing by the MOBH family relaxase TraI encoded within the gonococcal genetic island.
Nucleic Acids Res 47(15):8136-8153 (2019)
Heilers JH, Reiners J, Heller EM, Golzer A, Smits SHJ, van der Does C
|
| RgGuinier |
7.3 |
nm |
| Dmax |
31.4 |
nm |
| VolumePorod |
293 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Transcription intermediary factor 1-beta dimer, 82 kDa Mus musculus protein
|
| Buffer: |
10 mM Tris 300 mM NaCl 0.1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Aug 10
|
A Dissection of Oligomerisation by the TRIM28 Tripartite Motif and the Interaction with Members of the Krab-ZFP Family.
J Mol Biol (2019)
Sun Y, Keown JR, Black MM, Raclot C, Demarais N, Trono D, Turelli P, Goldstone DC
|
| RgGuinier |
7.0 |
nm |
| Dmax |
23.2 |
nm |
| VolumePorod |
232 |
nm3 |
|
|
|
|
|
|
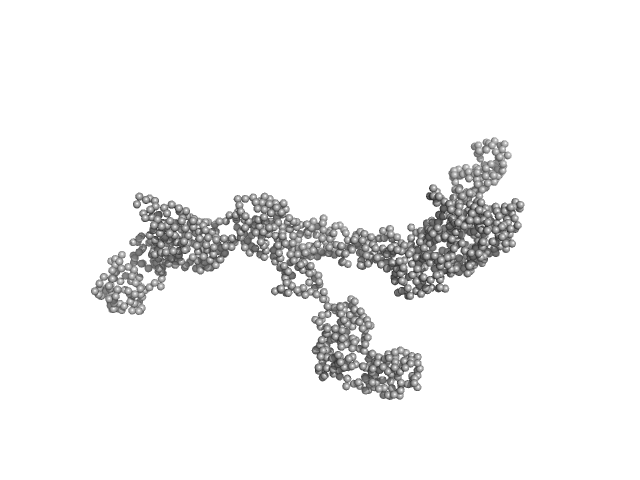
|
| Sample: |
Transcription intermediary factor 1-beta dimer, 82 kDa Mus musculus protein
Zinc finger protein 809 N-terminal MBP fusion monomer, 52 kDa Mus musculus protein
|
| Buffer: |
10 mM Tris 300 mM NaCl 0.1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Aug 10
|
A Dissection of Oligomerisation by the TRIM28 Tripartite Motif and the Interaction with Members of the Krab-ZFP Family.
J Mol Biol (2019)
Sun Y, Keown JR, Black MM, Raclot C, Demarais N, Trono D, Turelli P, Goldstone DC
|
| RgGuinier |
6.4 |
nm |
| Dmax |
22.0 |
nm |
| VolumePorod |
252 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
RoX2 stem-loop 7, 18-mer fragment monomer, 12 kDa synthetic construct RNA
|
| Buffer: |
20 mM NaPO4, 200 mM NaCl, 1 mM DTT, pH: 6.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Nov 29
|
Structure, dynamics and roX2-lncRNA binding of tandem double-stranded RNA binding domains dsRBD1,2 of Drosophila helicase Maleless.
Nucleic Acids Res 47(8):4319-4333 (2019)
Ankush Jagtap PK, Müller M, Masiewicz P, von Bülow S, Hollmann NM, Chen PC, Simon B, Thomae AW, Becker PB, Hennig J
|
| RgGuinier |
1.8 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
14 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Urokinase plasminogen activator surface receptor monomer, 37 kDa Homo sapiens protein
Urokinase-type plasminogen activator (Amino Terminal Fragment) monomer, 16 kDa Homo sapiens protein
|
| Buffer: |
20 mM PBS, 5 %(v/v) glycerol, 50 mM NaSO4,, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Jun 18
|
Did evolution create a flexible ligand-binding cavity in the urokinase receptor through deletion of a plesiotypic disulfide bond?
J Biol Chem (2019)
Leth JM, Mertens HDT, Leth-Espensen KZ, Jørgensen TJD, Ploug M
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.2 |
nm |
| VolumePorod |
102 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Urokinase plasminogen activator surface receptor monomer, 37 kDa Homo sapiens protein
|
| Buffer: |
20 mM PBS, 5 %(v/v) glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Dec 1
|
Did evolution create a flexible ligand-binding cavity in the urokinase receptor through deletion of a plesiotypic disulfide bond?
J Biol Chem (2019)
Leth JM, Mertens HDT, Leth-Espensen KZ, Jørgensen TJD, Ploug M
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.9 |
nm |
| VolumePorod |
55 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Urokinase plasminogen activator surface receptor monomer, 37 kDa Homo sapiens protein
Urokinase-type plasminogen activator (Amino Terminal Fragment) monomer, 16 kDa Homo sapiens protein
|
| Buffer: |
20 mM PBS, 5 %(v/v) glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Dec 1
|
Did evolution create a flexible ligand-binding cavity in the urokinase receptor through deletion of a plesiotypic disulfide bond?
J Biol Chem (2019)
Leth JM, Mertens HDT, Leth-Espensen KZ, Jørgensen TJD, Ploug M
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
77 |
nm3 |
|
|