|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDG25_fit1_model1.png)
|
| Sample: |
DNA repair protein RAD5 monomer, 138 kDa Saccharomyces cerevisiae protein
|
| Buffer: |
40 mM Tris, 150 mM KCl, 5 mM DTT, 5% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2019 Mar 6
|
Conformational flexibility of fork-remodeling helicase Rad5 shown by full-ensemble hybrid methods.
PLoS One 14(10):e0223875 (2019)
Gildenberg MS, Washington MT
|
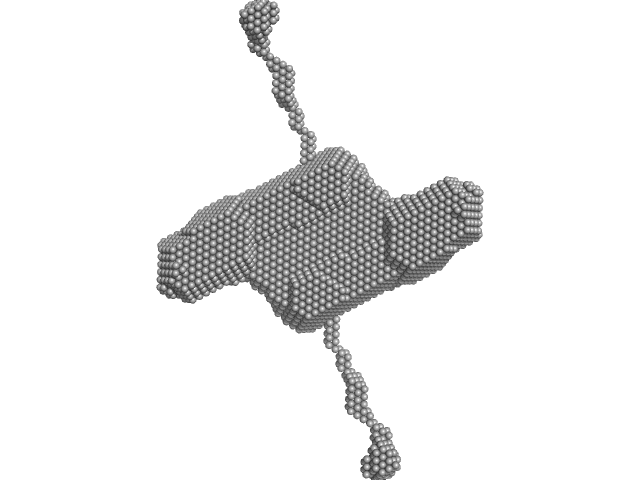
| RgGuinier |
4.7 |
nm |
| Dmax |
17.8 |
nm |
| VolumePorod |
254 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Lipoprotein lipase dimer, 101 kDa Homo sapiens protein
Glycosylphosphatidylinositol-anchored high density lipoprotein-binding protein 1 dimer, 29 kDa Homo sapiens protein
|
| Buffer: |
10 mM BisTris, 150 mM NaCl, 4 mM CaCl2, 10% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Dec 2
|
Structure of the lipoprotein lipase-GPIHBP1 complex that mediates plasma triglyceride hydrolysis.
Proc Natl Acad Sci U S A (2018)
Birrane G, Beigneux AP, Dwyer B, Strack-Logue B, Kristensen KK, Francone OL, Fong LG, Mertens HDT, Pan CQ, Ploug M, Young SG, Meiyappan M
|
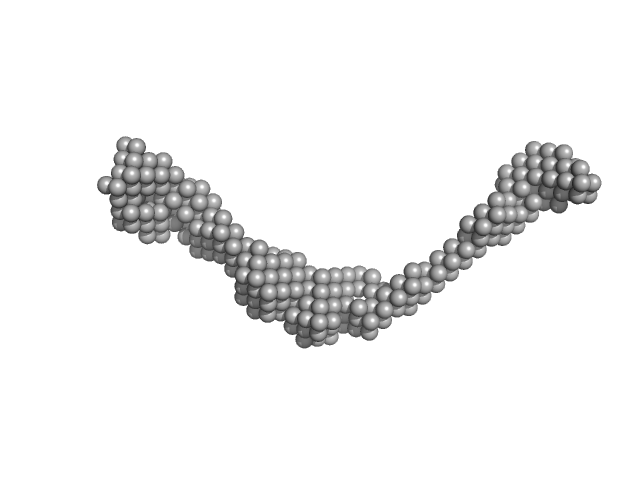
| RgGuinier |
5.5 |
nm |
| Dmax |
19.0 |
nm |
| VolumePorod |
319 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Bromodomain-containing protein 4 monomer, 47 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2% glycerol, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Jan 13
|
Interactome Rewiring Following Pharmacological Targeting of BET Bromodomains.
Mol Cell (2018)
Lambert JP, Picaud S, Fujisawa T, Hou H, Savitsky P, Uusküla-Reimand L, Gupta GD, Abdouni H, Lin ZY, Tucholska M, Knight JDR, Gonzalez-Badillo B, St-Denis N, Newman JA, Stucki M, Pelletier L, Bandeira N, Wilson MD, Filippakopoulos P, Gingras AC
|
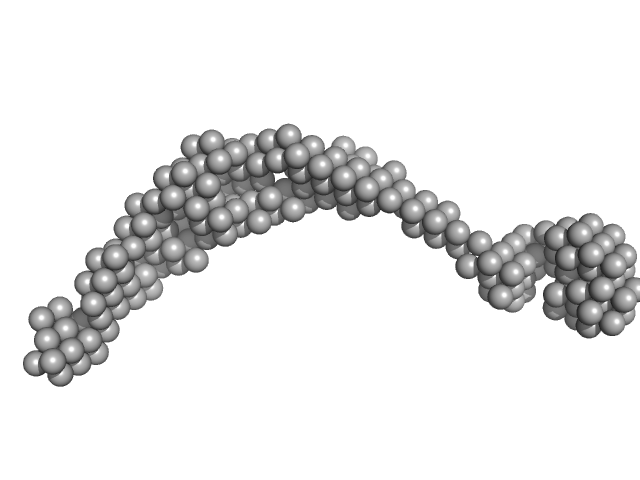
| RgGuinier |
6.3 |
nm |
| Dmax |
22.0 |
nm |
| VolumePorod |
251 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Bromodomain-containing protein 3 monomer, 44 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2% glycerol, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Jan 13
|
Interactome Rewiring Following Pharmacological Targeting of BET Bromodomains.
Mol Cell (2018)
Lambert JP, Picaud S, Fujisawa T, Hou H, Savitsky P, Uusküla-Reimand L, Gupta GD, Abdouni H, Lin ZY, Tucholska M, Knight JDR, Gonzalez-Badillo B, St-Denis N, Newman JA, Stucki M, Pelletier L, Bandeira N, Wilson MD, Filippakopoulos P, Gingras AC
|
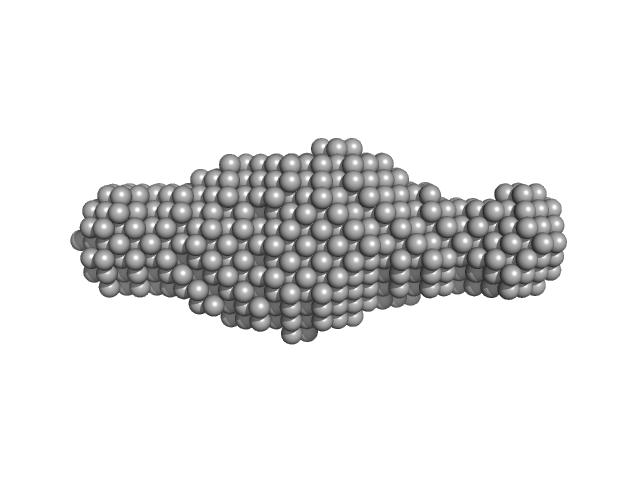
| RgGuinier |
6.2 |
nm |
| Dmax |
21.5 |
nm |
| VolumePorod |
210 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Bromodomain-containing protein 2 monomer, 43 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2% glycerol, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Jan 13
|
Interactome Rewiring Following Pharmacological Targeting of BET Bromodomains.
Mol Cell (2018)
Lambert JP, Picaud S, Fujisawa T, Hou H, Savitsky P, Uusküla-Reimand L, Gupta GD, Abdouni H, Lin ZY, Tucholska M, Knight JDR, Gonzalez-Badillo B, St-Denis N, Newman JA, Stucki M, Pelletier L, Bandeira N, Wilson MD, Filippakopoulos P, Gingras AC
|
| RgGuinier |
5.7 |
nm |
| Dmax |
21.0 |
nm |
| VolumePorod |
220 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Bromodomain testis-specific protein monomer, 43 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2% glycerol, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Jan 13
|
Interactome Rewiring Following Pharmacological Targeting of BET Bromodomains.
Mol Cell (2018)
Lambert JP, Picaud S, Fujisawa T, Hou H, Savitsky P, Uusküla-Reimand L, Gupta GD, Abdouni H, Lin ZY, Tucholska M, Knight JDR, Gonzalez-Badillo B, St-Denis N, Newman JA, Stucki M, Pelletier L, Bandeira N, Wilson MD, Filippakopoulos P, Gingras AC
|
| RgGuinier |
5.1 |
nm |
| Dmax |
20.0 |
nm |
| VolumePorod |
200 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Ribosome assembly protein 1 monomer, 124 kDa Saccharomyces cerevisiae protein
Ribosome maturation protein SDO1 monomer, 28 kDa Saccharomyces cerevisiae protein
|
| Buffer: |
50 mM Tris pH 8.0, 10% glycerol, 300 mM NaCl, 5 mM MgCl2., pH: |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Sep 21
|
Interaction of the GTPase Elongation Factor Like-1 with the Shwachman-Diamond Syndrome Protein and Its Missense Mutations.
Int J Mol Sci 19(12) (2018)
Gijsbers A, Montagut DC, Méndez-Godoy A, Altamura D, Saviano M, Siliqi D, Sánchez-Puig N
|
| RgGuinier |
5.0 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
333 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Proliferating cell nuclear antigen trimer, 117 kDa Saccharomyces cerevisiae protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 5% glycerol, 1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2018 Aug 6
|
Conformational Flexibility of Ubiquitin-Modified and SUMO-Modified PCNA Shown by Full-Ensemble Hybrid Methods.
J Mol Biol 430(24):5294-5303 (2018)
Powers KT, Lavering ED, Washington MT
|
| RgGuinier |
4.1 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
238 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Proliferating cell nuclear antigen trimer, 125 kDa Saccharomyces cerevisiae protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 5% glycerol, 1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2018 Aug 6
|
Conformational Flexibility of Ubiquitin-Modified and SUMO-Modified PCNA Shown by Full-Ensemble Hybrid Methods.
J Mol Biol 430(24):5294-5303 (2018)
Powers KT, Lavering ED, Washington MT
|
| RgGuinier |
4.7 |
nm |
| Dmax |
16.9 |
nm |
| VolumePorod |
310 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Old Yellow Enzyme of Leishmania braziliensis monomer, 42 kDa Leishmania braziliensis protein
|
| Buffer: |
25 mM Tris-HCl 100 mM NaCl and 1 mM β-mercaptoethanol, pH: 8 |
| Experiment: |
SAXS
data collected at SAXS1 Beamline, Brazilian Synchrotron Light Laboratory on 2015 May 26
|
Structural studies of Old Yellow Enzyme of Leishmania braziliensis in solution.
Arch Biochem Biophys (2018)
Walbert Veloso-Silva LL, Dores-Silva PR, Bertolino Reis DE, Moreno Oliveira LF, Libardi SH, Borges JC
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
73 |
nm3 |
|
|