|
|
|
|
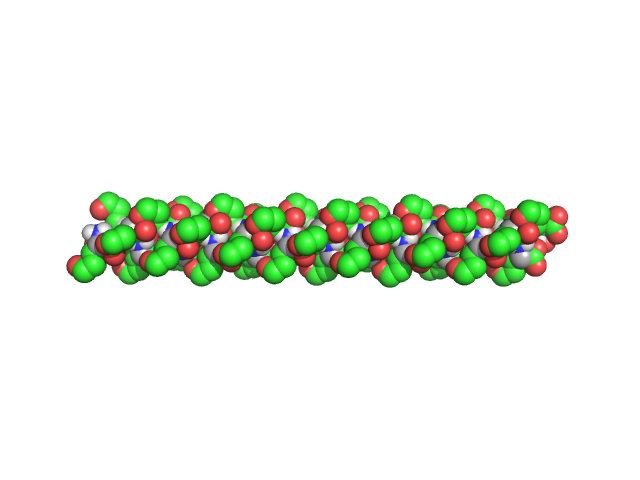
|
| Sample: |
Poly-L-Glutamic Acid, 6 kDa
|
| Buffer: |
Dimethyl Sulfoxide (DMSO), pH: |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Dec 11
|
β2-Type Amyloidlike Fibrils of Poly-l-glutamic Acid Convert into Long, Highly Ordered Helices upon Dissolution in Dimethyl Sulfoxide.
J Phys Chem B 122(50):11895-11905 (2018)
Berbeć S, Dec R, Molodenskiy D, Wielgus-Kutrowska B, Johannessen C, Hernik-Magoń A, Tobias F, Bzowska A, Ścibisz G, Keiderling TA, Svergun D, Dzwolak W
|
| RgGuinier |
1.8 |
nm |
| Dmax |
7.8 |
nm |
| VolumePorod |
5 |
nm3 |
|
|
|
|
|
|
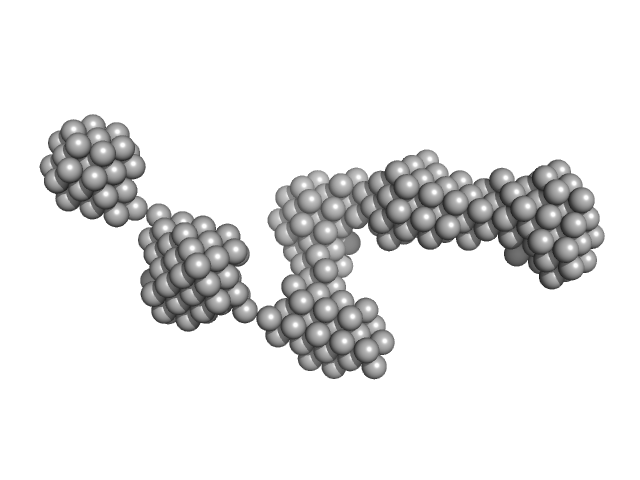
|
| Sample: |
Epstein-Barr nuclear antigen 2 dimer, 16 kDa Human gammaherpesvirus 4 protein
|
| Buffer: |
20mM Tris-HCl, 100mM NaCl, 2% Sucrose and 1mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Sep 23
|
Increased association between Epstein-Barr virus EBNA2 from type 2 strains and the transcriptional repressor BS69 restricts B cell growth
(2018)
Ponnusamy R, Khatri R, Correia P, Mancini E, Farrell P, West M
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.3 |
nm |
| VolumePorod |
19 |
nm3 |
|
|
|
|
|
|
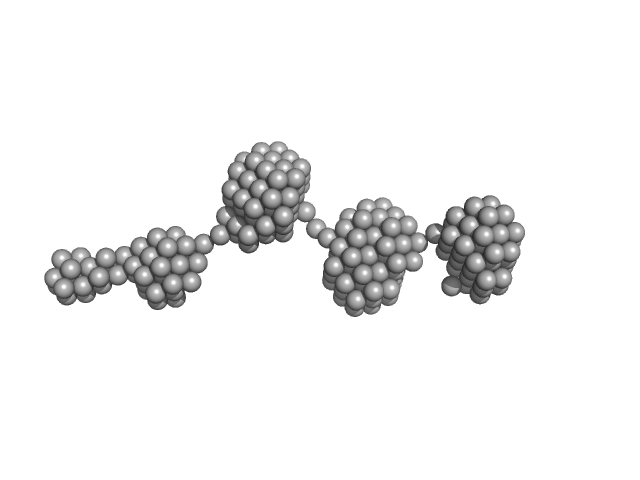
|
| Sample: |
Epstein-Barr nuclear antigen 2 dimer, 16 kDa Human gammaherpesvirus 4 protein
|
| Buffer: |
20mM Tris-HCl, 100mM NaCl, 2% Sucrose and 1mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Sep 23
|
Increased association between Epstein-Barr virus EBNA2 from type 2 strains and the transcriptional repressor BS69 restricts B cell growth
(2018)
Ponnusamy R, Khatri R, Correia P, Mancini E, Farrell P, West M
|
| RgGuinier |
2.8 |
nm |
| Dmax |
10.4 |
nm |
| VolumePorod |
20 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Zinc finger MYND domain-containing protein 11 tetramer, 60 kDa Homo sapiens protein
|
| Buffer: |
20mM Tris-HCl, 100mM NaCl, 2% Sucrose and 1mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Sep 23
|
Increased association between Epstein-Barr virus EBNA2 from type 2 strains and the transcriptional repressor BS69 restricts B cell growth
(2018)
Ponnusamy R, Khatri R, Correia P, Mancini E, Farrell P, West M
|
| RgGuinier |
5.3 |
nm |
| Dmax |
16.9 |
nm |
| VolumePorod |
83 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Epstein-Barr nuclear antigen 2 dimer, 16 kDa Human gammaherpesvirus 4 protein
Zinc finger MYND domain-containing protein 11 tetramer, 60 kDa Homo sapiens protein
|
| Buffer: |
20mM Tris-HCl, 100mM NaCl, 2% Sucrose and 1mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Nov 29
|
Increased association between Epstein-Barr virus EBNA2 from type 2 strains and the transcriptional repressor BS69 restricts B cell growth
(2018)
Ponnusamy R, Khatri R, Correia P, Mancini E, Farrell P, West M
|
| RgGuinier |
4.6 |
nm |
| Dmax |
13.8 |
nm |
| VolumePorod |
72 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Epstein-Barr nuclear antigen 2 dimer, 16 kDa Human gammaherpesvirus 4 protein
Zinc finger MYND domain-containing protein 11 hexamer, 90 kDa Homo sapiens protein
|
| Buffer: |
20mM Tris-HCl, 100mM NaCl, 2% Sucrose and 1mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Sep 23
|
Increased association between Epstein-Barr virus EBNA2 from type 2 strains and the transcriptional repressor BS69 restricts B cell growth
(2018)
Ponnusamy R, Khatri R, Correia P, Mancini E, Farrell P, West M
|
| RgGuinier |
4.7 |
nm |
| Dmax |
14.5 |
nm |
| VolumePorod |
239 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
DNA-(adenine N6)-methyltransferase monomer, 49 kDa Acinetobacter baumannii protein
|
| Buffer: |
150mM NaCl, 10mM Tris, 1mM DTT, 5% v/v glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Sep 30
|
Recombinant production of A1S_0222 from Acinetobacter baumannii ATCC 17978 and confirmation of its DNA-(adenine N6)-methyltransferase activity.
Protein Expr Purif 151:78-85 (2018)
Blaschke U, Suwono B, Zafari S, Ebersberger I, Skiebe E, Jeffries CM, Svergun DI, Wilharm G
|
| RgGuinier |
2.9 |
nm |
| Dmax |
11.1 |
nm |
| VolumePorod |
96 |
nm3 |
|
|
|
|
|
|
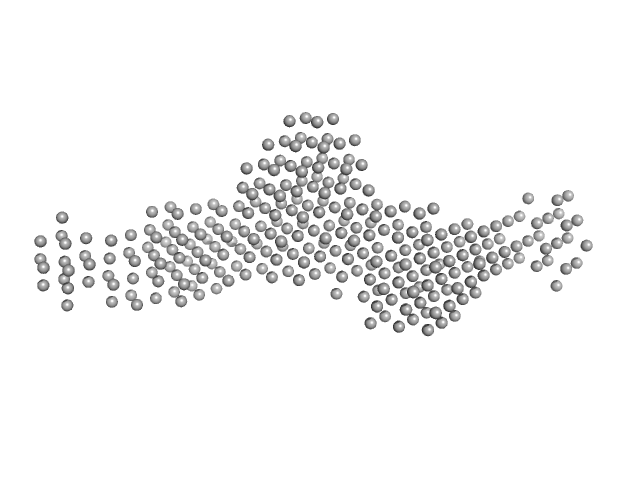
|
| Sample: |
SGT protein dimer, 92 kDa Leishmania braziliensis protein
|
| Buffer: |
20 mM Potassium Phosphate, 100 mM KCl, 10 mM EDTA, 1 mM B-mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS2 Beamline, Brazilian Synchrotron Light Laboratory on 2014 May 14
|
Structural and functional studies of the Leishmania braziliensis SGT co-chaperone indicate that it shares structural features with HIP and can interact with both Hsp90 and Hsp70 with similar affinities.
Int J Biol Macromol 118(Pt A):693-706 (2018)
Coto ALS, Seraphim TV, Batista FAH, Dores-Silva PR, Barranco ABF, Teixeira FR, Gava LM, Borges JC
|
| RgGuinier |
4.5 |
nm |
| Dmax |
17.0 |
nm |
|
|
|
|
|
|

|
| Sample: |
Calbindin monomer, 30 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150mM NaCl, pH: 7.8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Feb 28
|
The X-ray structure of human calbindin-D28K: an improved model.
Acta Crystallogr D Struct Biol 74(Pt 10):1008-1014 (2018)
Noble JW, Almalki R, Roe SM, Wagner A, Duman R, Atack JR
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
46 |
nm3 |
|
|
|
|
|
|
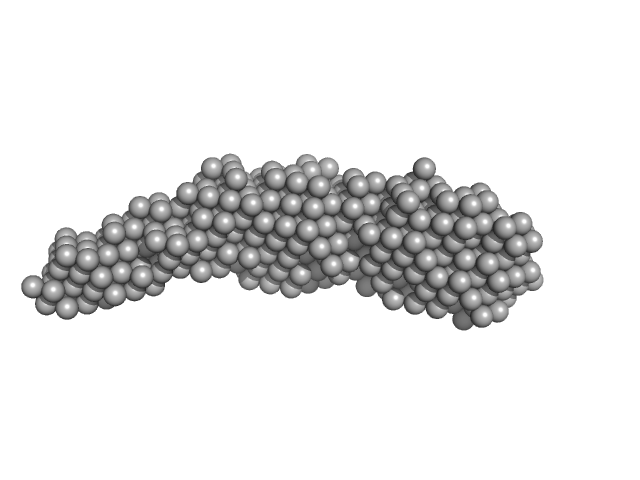
|
| Sample: |
ColH protein monomer, 24 kDa Hathewaya histolytica protein
Collagenous Peptide model [(PPG)10] trimer, 9 kDa synthetic construct protein
|
| Buffer: |
50 mM HEPES, 100 mM NaCl, 5 mM CaCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Oct 12
|
Ca2+ -induced orientation of tandem collagen binding domains from clostridial collagenase ColG permits two opposing functions of collagen fibril formation and retardation.
FEBS J 285(17):3254-3269 (2018)
Caviness P, Bauer R, Tanaka K, Janowska K, Roeser JR, Harter D, Sanders J, Ruth C, Matsushita O, Sakon J
|
| RgGuinier |
2.7 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
28 |
nm3 |
|
|