|
|
|
|
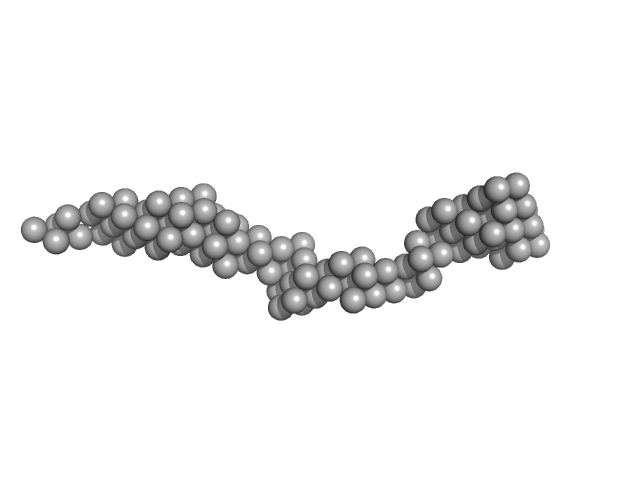
|
| Sample: |
Octo-repeat PrP mRNA mutant dimer, 144 kDa human PrP ORF RNA
|
| Buffer: |
10 mM Tris buffer with 100 mM LiCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Jun 6
|
Octa-repeat domain of the mammalian prion protein mRNA forms stable A-helical hairpin structure rather than G-quadruplexes.
Sci Rep 9(1):2465 (2019)
Czech A, Konarev PV, Goebel I, Svergun DI, Wills PR, Ignatova Z
|
| RgGuinier |
8.9 |
nm |
| Dmax |
33.0 |
nm |
| VolumePorod |
223 |
nm3 |
|
|
|
|
|
|
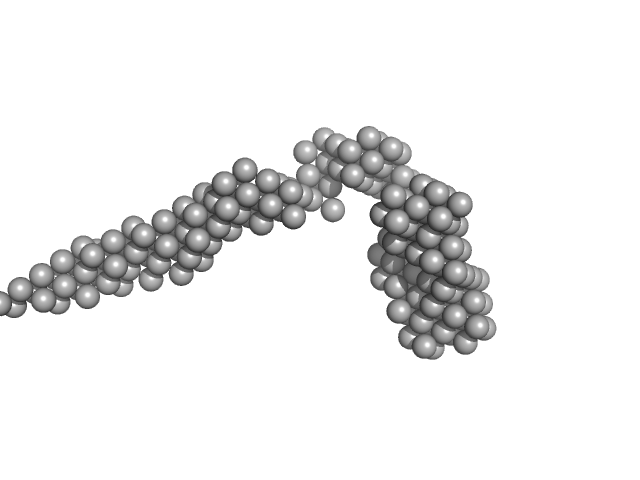
|
| Sample: |
Octo-repeat PrP mRNA mutant dimer, 144 kDa human PrP ORF RNA
|
| Buffer: |
10 mM Tris buffer with 100 mM KCl and 1 mM PDS, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Jun 6
|
Octa-repeat domain of the mammalian prion protein mRNA forms stable A-helical hairpin structure rather than G-quadruplexes.
Sci Rep 9(1):2465 (2019)
Czech A, Konarev PV, Goebel I, Svergun DI, Wills PR, Ignatova Z
|
| RgGuinier |
9.2 |
nm |
| Dmax |
33.0 |
nm |
| VolumePorod |
260 |
nm3 |
|
|
|
|
|
|
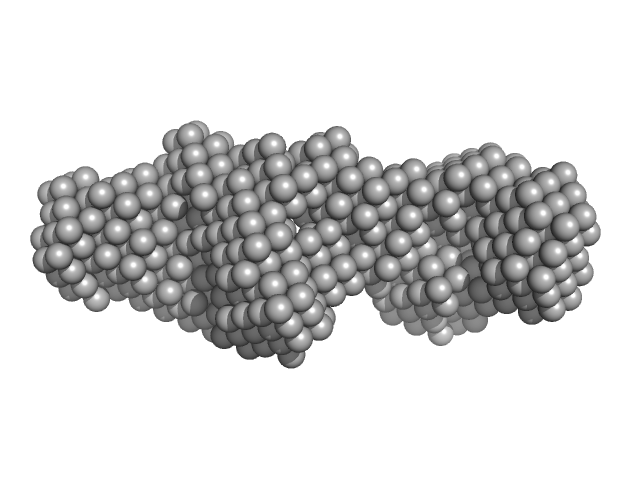
|
| Sample: |
LIM/homeobox protein Lhx4 monomer, 15 kDa Mus musculus protein
Insulin gene enhancer protein ISL-2 monomer, 4 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2015 Nov 19
|
Mutation in a flexible linker modulates binding affinity for modular complexes.
Proteins (2019)
Stokes PH, Robertson NO, Silva AP, Estephan T, Trewhella J, Guss JM, Matthews JM
|
| RgGuinier |
2.2 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
20 |
nm3 |
|
|
|
|
|
|
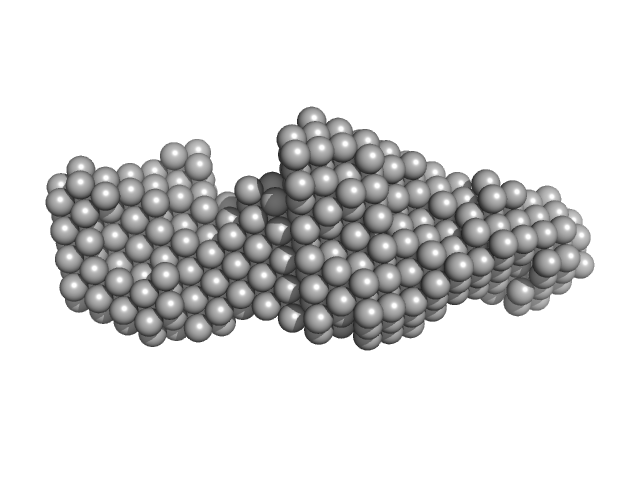
|
| Sample: |
LIM/homeobox protein Lhx4 monomer, 15 kDa Mus musculus protein
Insulin gene enhancer protein ISL-2 (R282G) monomer, 4 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2015 Nov 19
|
Mutation in a flexible linker modulates binding affinity for modular complexes.
Proteins (2019)
Stokes PH, Robertson NO, Silva AP, Estephan T, Trewhella J, Guss JM, Matthews JM
|
| RgGuinier |
2.3 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
21 |
nm3 |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDQP6_fit1_model1.png)
|
| Sample: |
Circularized Membrane scaffolding protein 1 E3 D1, 62 kDa protein
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) None, lipid
|
| Buffer: |
20 mM Tris-HCl pH 7.5, 100 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 May 5
|
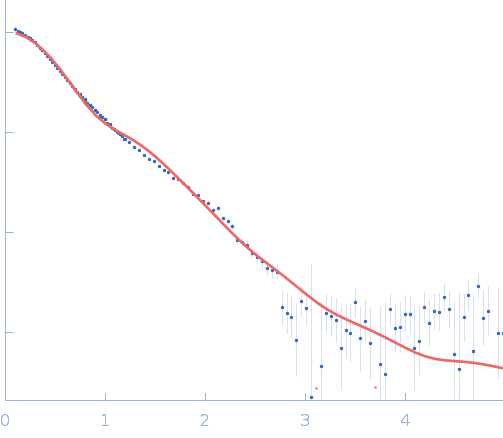
Circularized and solubility‐enhanced MSP
s facilitate simple and high‐yield production of stable nanodiscs for studies of membrane proteins in solution
The FEBS Journal 286(9):1734-1751 (2019)
Johansen N, Tidemand F, Nguyen T, Rand K, Pedersen M, Arleth L
|
| RgGuinier |
5.8 |
nm |
| Dmax |
14.5 |
nm |
|
|
|
|
|
|
|
| Sample: |
Sulfite reductase [NADPH] flavoprotein alpha-component (Assimilatory NADPH-dependent sulfite reductase flavoprotein) monomer, 61 kDa Escherichia coli (strain … protein
|
| Buffer: |
50 mM KPi, 100 mM NaCl, 1 mM EDTA, pH: 7.8 |
| Experiment: |
SANS
data collected at EQ-SANS (BL-6), Spallation Neutron Source on 2018 Jul 11
|
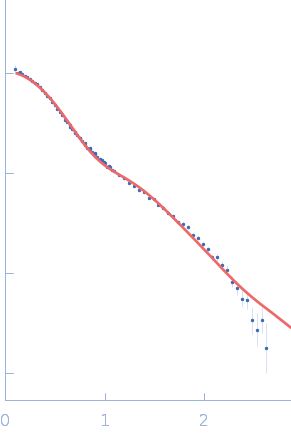
NADPH-dependent sulfite reductase flavoprotein adopts an extended conformation unique to this diflavin reductase
Journal of Structural Biology 205(2):170-179 (2019)
Tavolieri A, Murray D, Askenasy I, Pennington J, McGarry L, Stanley C, Stroupe M
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.6 |
nm |
| VolumePorod |
60 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Sulfite reductase [NADPH] flavoprotein alpha-component (Assimilatory NADPH-dependent sulfite reductase flavoprotein) monomer, 61 kDa Escherichia coli (strain … protein
|
| Buffer: |
50 mM KPi, 100 mM NaCl, 1 mM EDTA, pH: 7.8 |
| Experiment: |
SANS
data collected at EQ-SANS (BL-6), Spallation Neutron Source on 2018 Jul 11
|
NADPH-dependent sulfite reductase flavoprotein adopts an extended conformation unique to this diflavin reductase
Journal of Structural Biology 205(2):170-179 (2019)
Tavolieri A, Murray D, Askenasy I, Pennington J, McGarry L, Stanley C, Stroupe M
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.3 |
nm |
| VolumePorod |
73 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Diadenylate cyclase dimer, 39 kDa Staphylococcus aureus protein
Phosphoglucosamine mutase dimer, 99 kDa Staphylococcus aureus protein
|
| Buffer: |
30 mM Tris, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 May 7
|
Inhibition of the Staphylococcus aureus c-di-AMP cyclase DacA by direct interaction with the phosphoglucosamine mutase GlmM.
PLoS Pathog 15(1):e1007537 (2019)
Tosi T, Hoshiga F, Millership C, Singh R, Eldrid C, Patin D, Mengin-Lecreulx D, Thalassinos K, Freemont P, Gründling A
|
| RgGuinier |
3.9 |
nm |
| Dmax |
12.1 |
nm |
| VolumePorod |
204 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Phosphoglucosamine mutase dimer, 99 kDa Staphylococcus aureus protein
|
| Buffer: |
30 mM Tris, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 May 7
|
Inhibition of the Staphylococcus aureus c-di-AMP cyclase DacA by direct interaction with the phosphoglucosamine mutase GlmM.
PLoS Pathog 15(1):e1007537 (2019)
Tosi T, Hoshiga F, Millership C, Singh R, Eldrid C, Patin D, Mengin-Lecreulx D, Thalassinos K, Freemont P, Gründling A
|
| RgGuinier |
3.7 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
134 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Diadenylate cyclase dimer, 39 kDa Staphylococcus aureus protein
|
| Buffer: |
30 mM Tris, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 May 7
|
Inhibition of the Staphylococcus aureus c-di-AMP cyclase DacA by direct interaction with the phosphoglucosamine mutase GlmM.
PLoS Pathog 15(1):e1007537 (2019)
Tosi T, Hoshiga F, Millership C, Singh R, Eldrid C, Patin D, Mengin-Lecreulx D, Thalassinos K, Freemont P, Gründling A
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.6 |
nm |
| VolumePorod |
57 |
nm3 |
|
|