|
|
|
|

|
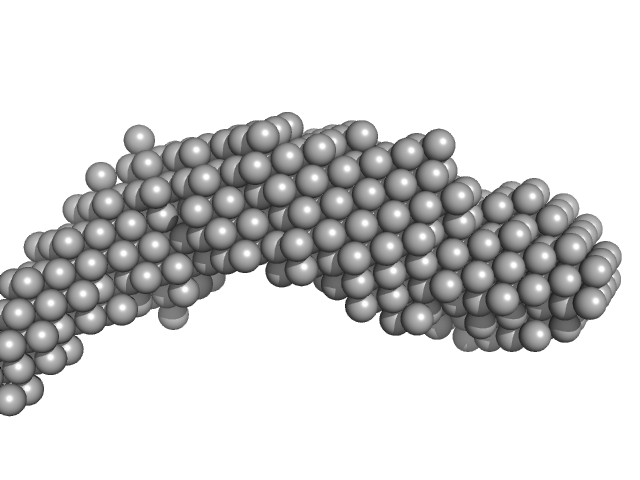
| Sample: |
E14ABC monomer, 106 kDa DNA
|
| Buffer: |
154 mM NaCl, pH: 8.3 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Jun 16
|
Optical and Structural Characterization of a Chronic Myeloid Leukemia DNA Biosensor.
ACS Chem Biol 13(5):1235-1242 (2018)
Cordeiro M, Otrelo-Cardoso AR, Svergun DI, Konarev PV, Lima JC, Santos-Silva T, Baptista PV
|
| RgGuinier |
4.7 |
nm |
| Dmax |
25.0 |
nm |
| VolumePorod |
152 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
E13A monomer, 17 kDa DNA
|
| Buffer: |
154 mM NaCl, pH: 8.3 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2013 Nov 22
|
Optical and Structural Characterization of a Chronic Myeloid Leukemia DNA Biosensor.
ACS Chem Biol 13(5):1235-1242 (2018)
Cordeiro M, Otrelo-Cardoso AR, Svergun DI, Konarev PV, Lima JC, Santos-Silva T, Baptista PV
|
| RgGuinier |
2.3 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
24 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
E13B monomer, 10 kDa DNA
|
| Buffer: |
154 mM NaCl, pH: 8.3 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2013 Nov 22
|
Optical and Structural Characterization of a Chronic Myeloid Leukemia DNA Biosensor.
ACS Chem Biol 13(5):1235-1242 (2018)
Cordeiro M, Otrelo-Cardoso AR, Svergun DI, Konarev PV, Lima JC, Santos-Silva T, Baptista PV
|
| RgGuinier |
1.8 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
13 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
E13C monomer, 6 kDa DNA
|
| Buffer: |
154 mM NaCl, pH: 8.3 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2013 Nov 22
|
Optical and Structural Characterization of a Chronic Myeloid Leukemia DNA Biosensor.
ACS Chem Biol 13(5):1235-1242 (2018)
Cordeiro M, Otrelo-Cardoso AR, Svergun DI, Konarev PV, Lima JC, Santos-Silva T, Baptista PV
|
| RgGuinier |
1.3 |
nm |
| Dmax |
5.0 |
nm |
| VolumePorod |
8 |
nm3 |
|
|
|
|
|
|
|
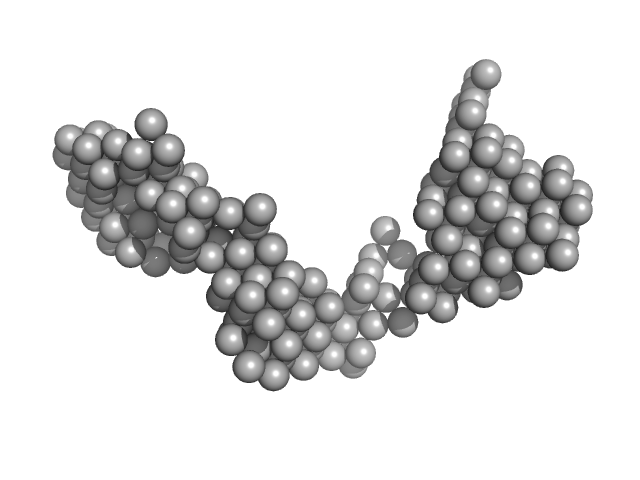
| Sample: |
E13AB monomer, 79 kDa DNA
|
| Buffer: |
154 mM NaCl, pH: 8.3 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2013 Nov 22
|
Optical and Structural Characterization of a Chronic Myeloid Leukemia DNA Biosensor.
ACS Chem Biol 13(5):1235-1242 (2018)
Cordeiro M, Otrelo-Cardoso AR, Svergun DI, Konarev PV, Lima JC, Santos-Silva T, Baptista PV
|
| RgGuinier |
4.7 |
nm |
| Dmax |
20.0 |
nm |
| VolumePorod |
103 |
nm3 |
|
|
|
|
|
|
|
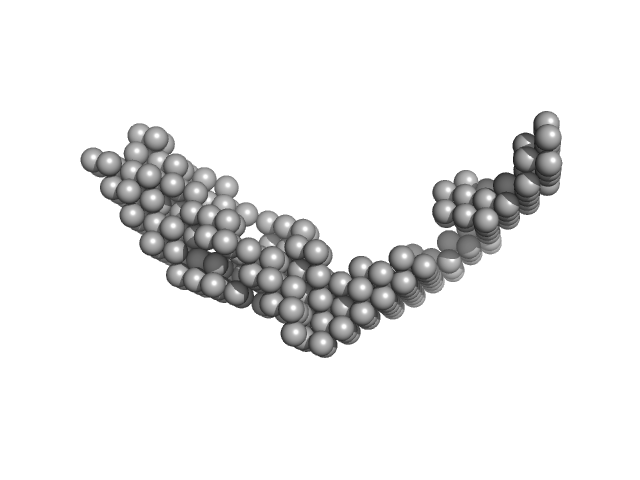
| Sample: |
E13ABC monomer, 42 kDa DNA
|
| Buffer: |
154 mM NaCl, pH: 8.3 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2013 Nov 22
|
Optical and Structural Characterization of a Chronic Myeloid Leukemia DNA Biosensor.
ACS Chem Biol 13(5):1235-1242 (2018)
Cordeiro M, Otrelo-Cardoso AR, Svergun DI, Konarev PV, Lima JC, Santos-Silva T, Baptista PV
|
| RgGuinier |
3.9 |
nm |
| Dmax |
18.0 |
nm |
| VolumePorod |
59 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
E13Ae14Be13C monomer, 40 kDa DNA
|
| Buffer: |
154 mM NaCl, pH: 8.3 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Jun 16
|
Optical and Structural Characterization of a Chronic Myeloid Leukemia DNA Biosensor.
ACS Chem Biol 13(5):1235-1242 (2018)
Cordeiro M, Otrelo-Cardoso AR, Svergun DI, Konarev PV, Lima JC, Santos-Silva T, Baptista PV
|
| RgGuinier |
3.7 |
nm |
| Dmax |
18.0 |
nm |
| VolumePorod |
65 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
E14Ae13Be14C monomer, 31 kDa DNA
|
| Buffer: |
154 mM NaCl, pH: 8.3 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Jun 16
|
Optical and Structural Characterization of a Chronic Myeloid Leukemia DNA Biosensor.
ACS Chem Biol 13(5):1235-1242 (2018)
Cordeiro M, Otrelo-Cardoso AR, Svergun DI, Konarev PV, Lima JC, Santos-Silva T, Baptista PV
|
| RgGuinier |
3.4 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
42 |
nm3 |
|
|
|
|
|
|
|
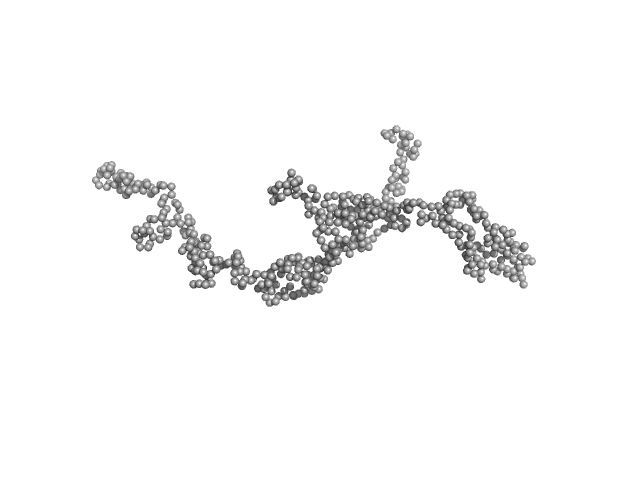
| Sample: |
Dystrophin central domain repeats 16 to 21 (Δ2146-2305; Becker muscular dystrophy variant, deletion of exons 45-47) monomer, 64 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris 150 mM NaCl 1 mM EDTA 2% glycerol 5% acetonitrile, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2014 Feb 5
|
Dystrophin's central domain forms a complex filament that becomes disorganized by in-frame deletions.
J Biol Chem 293(18):6637-6646 (2018)
Delalande O, Molza AE, Dos Santos Morais R, Chéron A, Pollet É, Raguenes-Nicol C, Tascon C, Giudice E, Guilbaud M, Nicolas A, Bondon A, Leturcq F, Férey N, Baaden M, Perez J, Roblin P, Piétri-Rouxel F, Hubert JF, Czjzek M, Le Rumeur E
|
| RgGuinier |
6.0 |
nm |
| Dmax |
21.0 |
nm |
| VolumePorod |
184 |
nm3 |
|
|
|
|
|
|
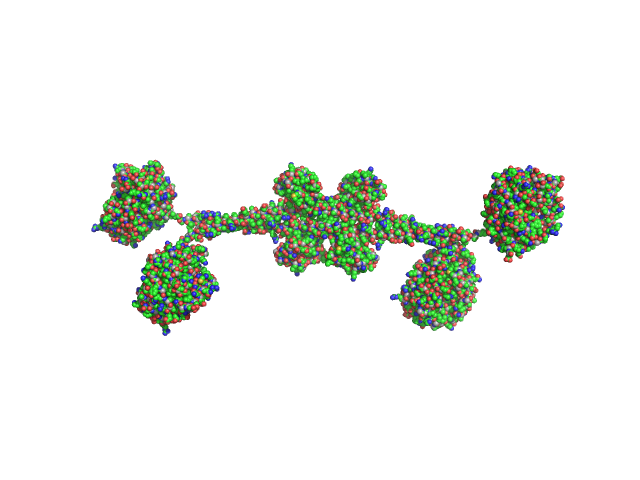
|
| Sample: |
Full-length hypothetical protein CTHT_0072540 tetramer, 207 kDa Chaetomium thermophilum protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 2 mM β-mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at I911-4, MAX IV on 2013 Oct 15
|
Prp19/Pso4 Is an Autoinhibited Ubiquitin Ligase Activated by Stepwise Assembly of Three Splicing Factors.
Mol Cell 69(6):979-992.e6 (2018)
de Moura TR, Mozaffari-Jovin S, Szabó CZK, Schmitzová J, Dybkov O, Cretu C, Kachala M, Svergun D, Urlaub H, Lührmann R, Pena V
|
| RgGuinier |
6.2 |
nm |
| Dmax |
23.0 |
nm |
| VolumePorod |
280 |
nm3 |
|
|