|
|
|
|

|
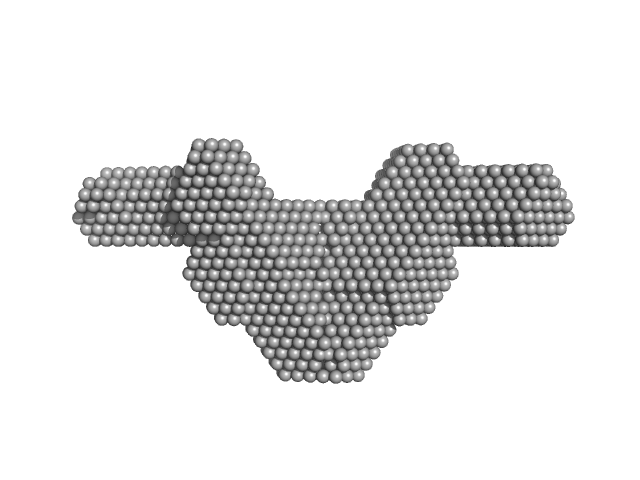
| Sample: |
Fc fragment of IgG binding protein dimer, 552 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES, 100 mM NaCl, 10 mM CaCl2, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2020 Nov 20
|
The structure of FCGBP is formed as a disulfide-mediated homodimer between its C-terminal domains.
FEBS J (2025)
Ehrencrona E, Gallego P, Trillo-Muyo S, Garcia-Bonete MJ, Recktenwald CV, Hansson GC, Johansson MEV
|
| RgGuinier |
10.0 |
nm |
| Dmax |
42.0 |
nm |
| VolumePorod |
1462 |
nm3 |
|
|
|
|
|
|
|
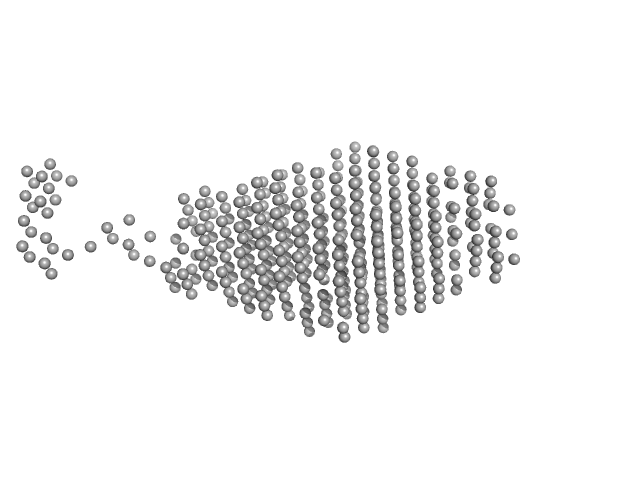
| Sample: |
Non-POU domain-containing octamer-binding protein dimer, 60 kDa Homo sapiens protein
Splicing factor, proline- and glutamine-rich SFPQ276-598(R542C)/NONO53-312 dimer) dimer, 76 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 250 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2015 Apr 28
|
Structural plasticity of the coiled-coil interactions in human SFPQ.
Nucleic Acids Res (2024)
Koning HJ, Lai JY, Marshall AC, Stroeher E, Monahan G, Pullakhandam A, Knott GJ, Ryan TM, Fox AH, Whitten A, Lee M, Bond CS
|
| RgGuinier |
5.2 |
nm |
| Dmax |
21.0 |
nm |
| VolumePorod |
257 |
nm3 |
|
|
|
|
|
|
|
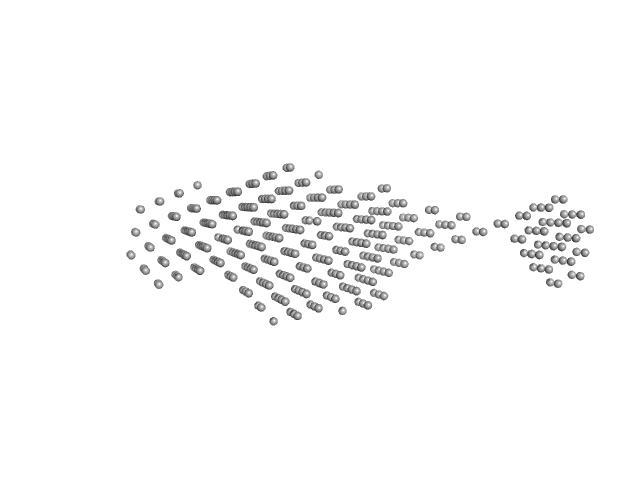
| Sample: |
Sensor domain-containing diguanylate cyclase dimer, 72 kDa Methylotenera sp. protein
|
| Buffer: |
10 mM Tris, 50 mM NaCl, 2 mM MgCl2, 3% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Apr 24
|
LOV-activated diguanylate cyclase
Ursula Vide
|
| RgGuinier |
3.4 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
128 |
nm3 |
|
|
|
|
|
|
|
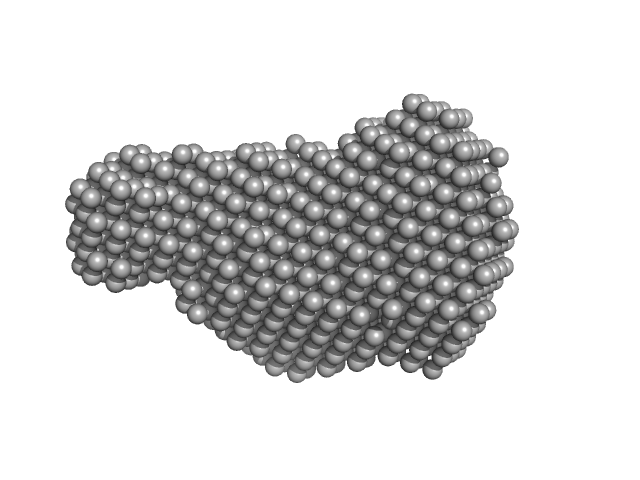
| Sample: |
Sensor domain-containing diguanylate cyclase dimer, 72 kDa Methylotenera sp. protein
|
| Buffer: |
10 mM Tris, 50 mM NaCl, 2 mM MgCl2, 3% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Apr 24
|
LOV-activated diguanylate cyclase
|
| RgGuinier |
5.2 |
nm |
| Dmax |
18.0 |
nm |
| VolumePorod |
159 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Virus termination factor small subunit monomer, 33 kDa Monkeypox virus (strain … protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2024 Jan 4
|
Structural basis of the monkeypox virus mRNA cap N7 methyltransferase complex.
Emerg Microbes Infect 13(1):2369193 (2024)
Chen A, Fang N, Zhang Z, Wen Y, Shen Y, Zhang Y, Zhang L, Zhao G, Ding J, Li J
|
| RgGuinier |
2.4 |
nm |
| Dmax |
6.8 |
nm |
| VolumePorod |
51 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Virus termination factor small subunit monomer, 33 kDa Monkeypox virus (strain … protein
MRNA-capping enzyme catalytic subunit monomer, 35 kDa Monkeypox virus (strain … protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2024 Jan 4
|
Structural basis of the monkeypox virus mRNA cap N7 methyltransferase complex.
Emerg Microbes Infect 13(1):2369193 (2024)
Chen A, Fang N, Zhang Z, Wen Y, Shen Y, Zhang Y, Zhang L, Zhao G, Ding J, Li J
|
| RgGuinier |
2.9 |
nm |
| Dmax |
10.1 |
nm |
| VolumePorod |
106 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Xylose isomerase tetramer, 196 kDa Bacteroides thetaiotaomicron (strain … protein
|
| Buffer: |
PBS, pH: 7.4 |
| Experiment: |
SAXS
data collected at BioSAXS, Australian Synchrotron on 2024 Oct 22
|
BioSAXS Australian Synchrotron Standard Protein
Annmaree Warrender
|
| RgGuinier |
3.5 |
nm |
| Dmax |
9.9 |
nm |
| VolumePorod |
222 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Ras-related protein Rab-1A monomer, 24 kDa Homo sapiens protein
|
| Buffer: |
300 mM NaCl, 2 mM 2-mercaptoethanol and 30 mM Tris-HCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2015 Nov 17
|
The SAXS Analysis of the human GTPase Rab1a
Wenhua Zhang
|
| RgGuinier |
2.4 |
nm |
| Dmax |
10.2 |
nm |
| VolumePorod |
36 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Uncharacterized protein SAUSA300_1119 dimer, 125 kDa Staphylococcus aureus (strain … protein
|
| Buffer: |
50 mM Tris, 150 mM KCl, 1 mM TCEP, 5% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Oct 5
|
Molecular insights into the structure and function of the Staphylococcus aureus fatty acid kinase.
J Biol Chem 300(12):107920 (2024)
Myers MJ, Xu Z, Ryan BJ, DeMars ZR, Ridder MJ, Johnson DK, Krute CN, Flynn TS, Kashipathy MM, Battaile KP, Schnicker N, Lovell S, Freudenthal BD, Bose JL
|
| RgGuinier |
4.3 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
219 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
G-quadrupex monomer, 6 kDa Hepatitis B virus DNA
|
| Buffer: |
20 mM HEPES, 100 mM KCl, 1 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Jan 15
|
G-quadruplex from HBV genome
Trushar Patel
|
| RgGuinier |
1.7 |
nm |
| Dmax |
4.0 |
nm |
| VolumePorod |
13 |
nm3 |
|
|