|
|
|
|
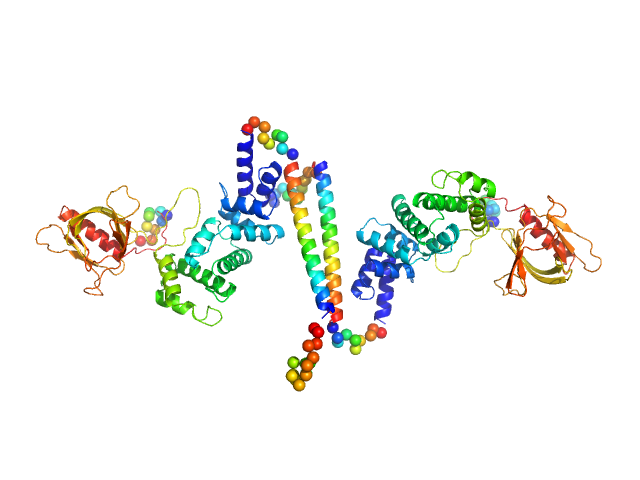
|
| Sample: |
Cytohesin-3 dimer, 90 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 2 mM MgCl2, 0.1% 2-mercaptoethanol, 5% glycerol, 0.001 mM insitol 1,3,4,5-tetrakis phosphate, pH: 8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2013 Nov 15
|
Structural Organization and Dynamics of Homodimeric Cytohesin Family Arf GTPase Exchange Factors in Solution and on Membranes.
Structure (2019)
Das S, Malaby AW, Nawrotek A, Zhang W, Zeghouf M, Maslen S, Skehel M, Chakravarthy S, Irving TC, Bilsel O, Cherfils J, Lambright DG
|
| RgGuinier |
5.1 |
nm |
| Dmax |
25.7 |
nm |
| VolumePorod |
168 |
nm3 |
|
|
|
|
|
|
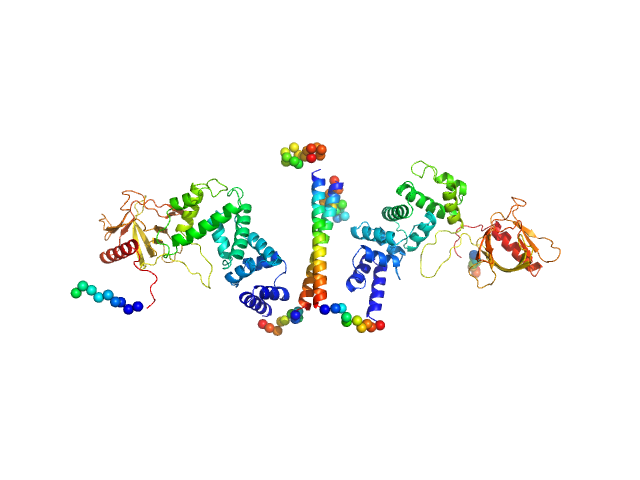
|
| Sample: |
Cytohesin-3 dimer, 90 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 2 mM MgCl2, 0.1% 2-mercaptoethanol, 5% glycerol, 0.001 mM insitol 1,3,4,5-tetrakis phosphate, pH: 8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2013 Nov 15
|
Structural Organization and Dynamics of Homodimeric Cytohesin Family Arf GTPase Exchange Factors in Solution and on Membranes.
Structure (2019)
Das S, Malaby AW, Nawrotek A, Zhang W, Zeghouf M, Maslen S, Skehel M, Chakravarthy S, Irving TC, Bilsel O, Cherfils J, Lambright DG
|
| RgGuinier |
5.1 |
nm |
| Dmax |
25.7 |
nm |
| VolumePorod |
168 |
nm3 |
|
|
|
|
|
|
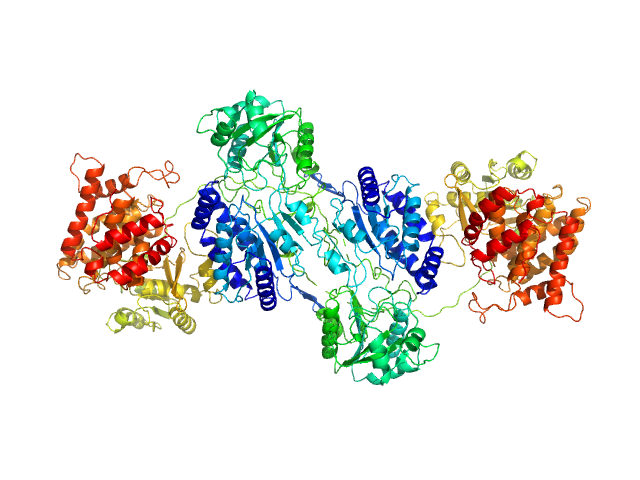
|
| Sample: |
F670E Aldehyde-alcohol dehydrogenase dimer, 192 kDa Escherichia coli protein
|
| Buffer: |
50 mM HEPES pH 7, 500 mM NaCl, 5% (v/v) glycerol, pH: 7 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Oct 4
|
Aldehyde-alcohol dehydrogenase forms a high-order spirosome architecture critical for its activity.
Nat Commun 10(1):4527 (2019)
Kim G, Azmi L, Jang S, Jung T, Hebert H, Roe AJ, Byron O, Song JJ
|
| RgGuinier |
5.0 |
nm |
| Dmax |
17.4 |
nm |
| VolumePorod |
260 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Zinc protease PqqL monomer, 102 kDa Escherichia coli protein
|
| Buffer: |
20 mM Tris HCl, 150 nM NaCl, 0.02 % NaN3, 5% glycerol, pH: 7.8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Jun 14
|
Protease-associated import systems are widespread in Gram-negative bacteria.
PLoS Genet 15(10):e1008435 (2019)
Grinter R, Leung PM, Wijeyewickrema LC, Littler D, Beckham S, Pike RN, Walker D, Greening C, Lithgow T
|
| RgGuinier |
4.0 |
nm |
| Dmax |
13.7 |
nm |
| VolumePorod |
207 |
nm3 |
|
|
|
|
|
|
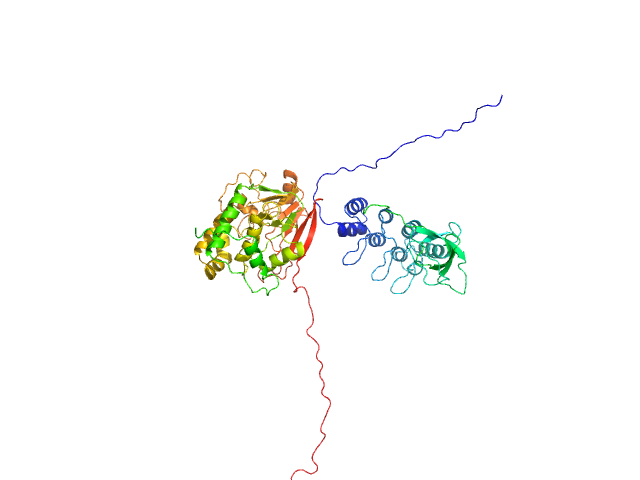
|
| Sample: |
Serine/threonine-protein phosphatase PP1-alpha catalytic subunit monomer, 38 kDa Homo sapiens protein
Inhibitor of apoptosis-stimulating protein of p53 (RelA-associated inhibitor) monomer, 25 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris, 150 mM NaCl, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Apr 25
|
Flexible Tethering of ASPP Proteins Facilitates PP-1c Catalysis.
Structure 27(10):1485-1496.e4 (2019)
Zhou Y, Millott R, Kim HJ, Peng S, Edwards RA, Skene-Arnold T, Hammel M, Lees-Miller SP, Tainer JA, Holmes CFB, Glover JNM
|
| RgGuinier |
3.4 |
nm |
| Dmax |
12.4 |
nm |
| VolumePorod |
132 |
nm3 |
|
|
|
|
|
|
|
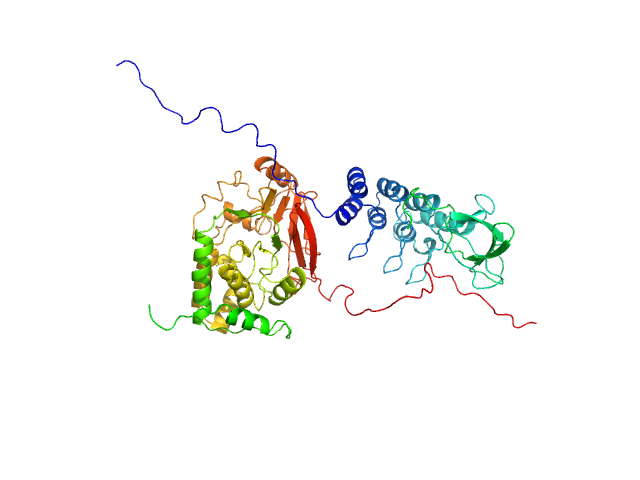
| Sample: |
Serine/threonine-protein phosphatase PP1-alpha catalytic subunit monomer, 38 kDa Homo sapiens protein
Inhibitor of apoptosis-stimulating protein of p53 (RelA-associated inhibitor) monomer, 25 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris, 150 mM NaCl, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Apr 25
|
Flexible Tethering of ASPP Proteins Facilitates PP-1c Catalysis.
Structure 27(10):1485-1496.e4 (2019)
Zhou Y, Millott R, Kim HJ, Peng S, Edwards RA, Skene-Arnold T, Hammel M, Lees-Miller SP, Tainer JA, Holmes CFB, Glover JNM
|
| RgGuinier |
3.0 |
nm |
| Dmax |
10.9 |
nm |
| VolumePorod |
116 |
nm3 |
|
|
|
|
|
|
|
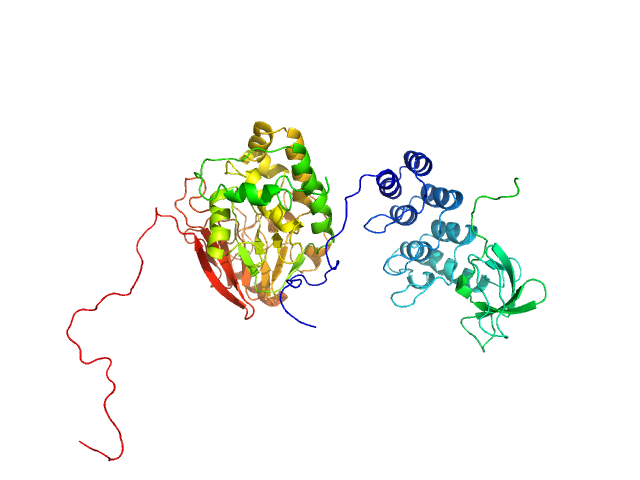
| Sample: |
Inhibitor of apoptosis-stimulating protein of p53 (RelA-associated inhibitor) monomer, 25 kDa Homo sapiens protein
Serine/threonine-protein phosphatase PP1-alpha catalytic subunit monomer, 38 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris, 150 mM NaCl, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 May 31
|
Flexible Tethering of ASPP Proteins Facilitates PP-1c Catalysis.
Structure 27(10):1485-1496.e4 (2019)
Zhou Y, Millott R, Kim HJ, Peng S, Edwards RA, Skene-Arnold T, Hammel M, Lees-Miller SP, Tainer JA, Holmes CFB, Glover JNM
|
| RgGuinier |
3.1 |
nm |
| Dmax |
11.3 |
nm |
| VolumePorod |
157 |
nm3 |
|
|
|
|
|
|
|
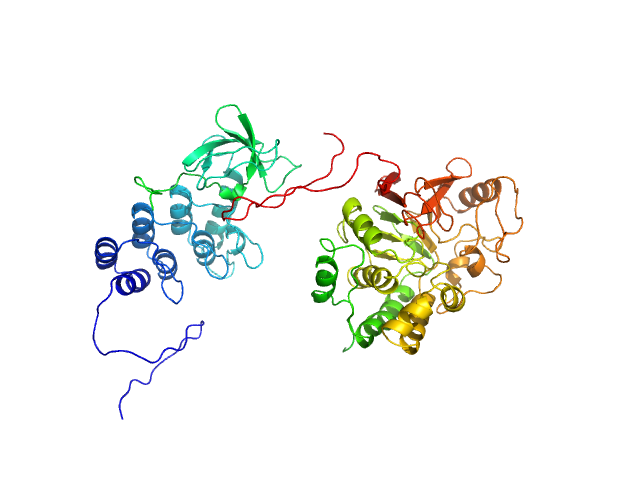
| Sample: |
Inhibitor of apoptosis-stimulating protein of p53 (RelA-associated inhibitor) monomer, 25 kDa Homo sapiens protein
Serine/threonine-protein phosphatase PP1-alpha catalytic subunit monomer, 38 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris, 150 mM NaCl, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 May 31
|
Flexible Tethering of ASPP Proteins Facilitates PP-1c Catalysis.
Structure 27(10):1485-1496.e4 (2019)
Zhou Y, Millott R, Kim HJ, Peng S, Edwards RA, Skene-Arnold T, Hammel M, Lees-Miller SP, Tainer JA, Holmes CFB, Glover JNM
|
| RgGuinier |
3.4 |
nm |
| Dmax |
12.2 |
nm |
| VolumePorod |
180 |
nm3 |
|
|
|
|
|
|
|
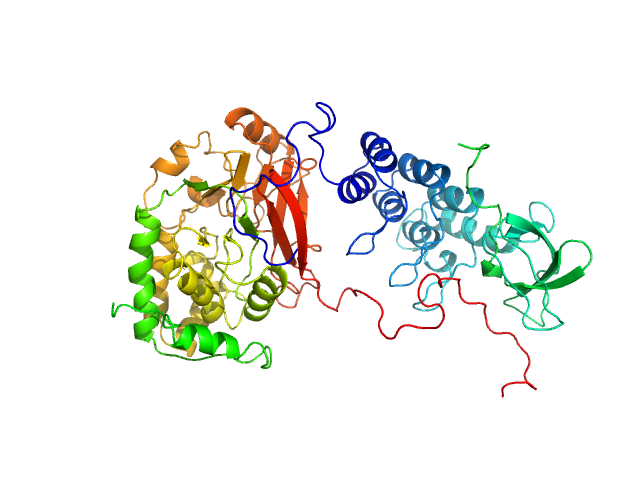
| Sample: |
Serine/threonine-protein phosphatase PP1-alpha catalytic subunit monomer, 38 kDa Homo sapiens protein
Apoptosis-stimulating of p53 protein 2 monomer, 26 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris, 150 mM NaCl, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 May 31
|
Flexible Tethering of ASPP Proteins Facilitates PP-1c Catalysis.
Structure 27(10):1485-1496.e4 (2019)
Zhou Y, Millott R, Kim HJ, Peng S, Edwards RA, Skene-Arnold T, Hammel M, Lees-Miller SP, Tainer JA, Holmes CFB, Glover JNM
|
| RgGuinier |
2.9 |
nm |
| Dmax |
10.2 |
nm |
| VolumePorod |
144 |
nm3 |
|
|
|
|
|
|
|
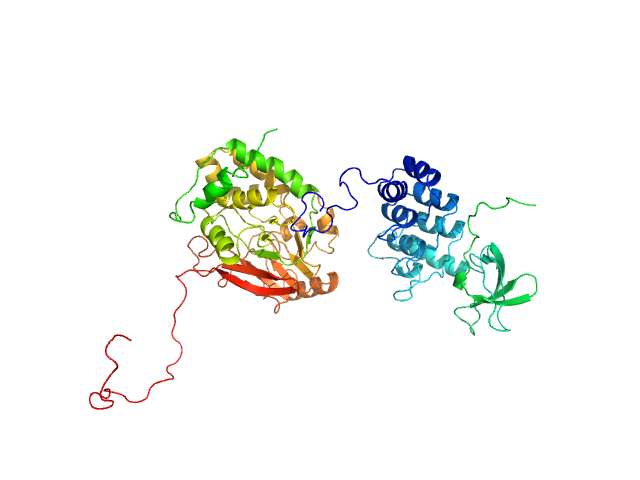
| Sample: |
Serine/threonine-protein phosphatase PP1-alpha catalytic subunit monomer, 38 kDa Homo sapiens protein
Apoptosis-stimulating of p53 protein 2 monomer, 26 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris, 150 mM NaCl, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 May 31
|
Flexible Tethering of ASPP Proteins Facilitates PP-1c Catalysis.
Structure 27(10):1485-1496.e4 (2019)
Zhou Y, Millott R, Kim HJ, Peng S, Edwards RA, Skene-Arnold T, Hammel M, Lees-Miller SP, Tainer JA, Holmes CFB, Glover JNM
|
| RgGuinier |
3.3 |
nm |
| Dmax |
11.7 |
nm |
| VolumePorod |
172 |
nm3 |
|
|