|
|
|
|
|
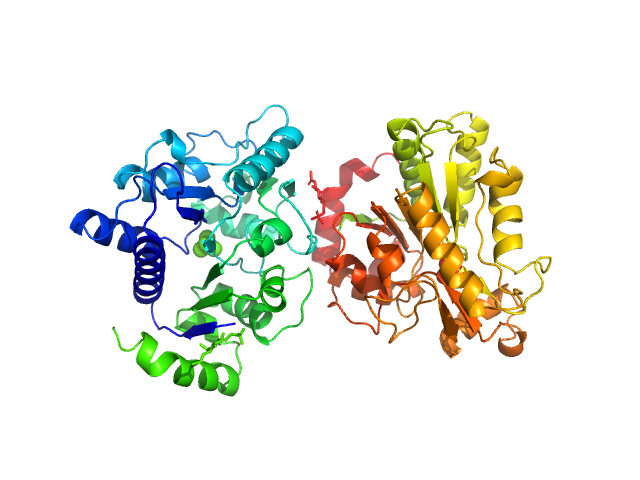
| Sample: |
Glucosamine-6-phosphate deaminase dimer, 57 kDa Staphylococcus aureus protein
|
| Buffer: |
20 mM Tris-HCl 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2016 Apr 26
|
Functional and solution structure studies of amino sugar deacetylase and deaminase enzymes from Staphylococcus aureus.
FEBS Lett (2018)
Davies JS, Coombes D, Horne CR, Pearce FG, Friemann R, North RA, Dobson RCJ
|
| RgGuinier |
2.7 |
nm |
| Dmax |
8.9 |
nm |
| VolumePorod |
84 |
nm3 |
|
|
|
|
|
|
|
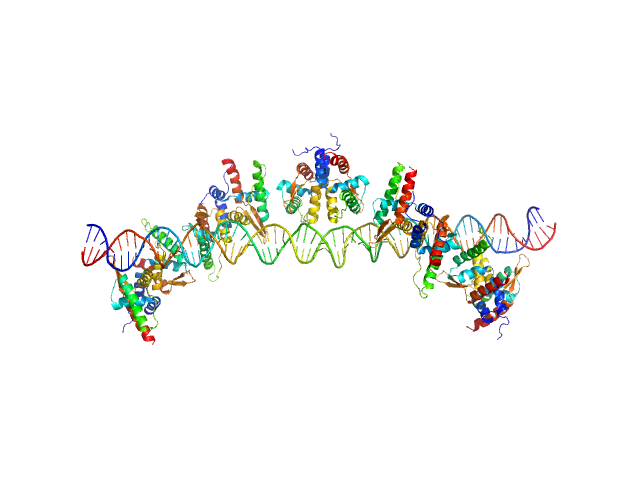
| Sample: |
S48 DNA strand 1 monomer, 21 kDa DNA
S48 DNA strand 2 monomer, 21 kDa DNA
TubR of the pXO1-like plasmid pBc10987 from B. cereus (Bc-TubR) decamer, 137 kDa protein
|
| Buffer: |
0.1 M NaCl, 10 mM Tris, pH: 8 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2017 Nov 28
|
Cooperative DNA Binding of the Plasmid Partitioning Protein TubR from the Bacillus cereus pXO1 Plasmid.
J Mol Biol (2018)
Hayashi I, Oda T, Sato M, Fuchigami S
|
| RgGuinier |
6.1 |
nm |
| Dmax |
23.0 |
nm |
| VolumePorod |
305 |
nm3 |
|
|
|
|
|
|
|
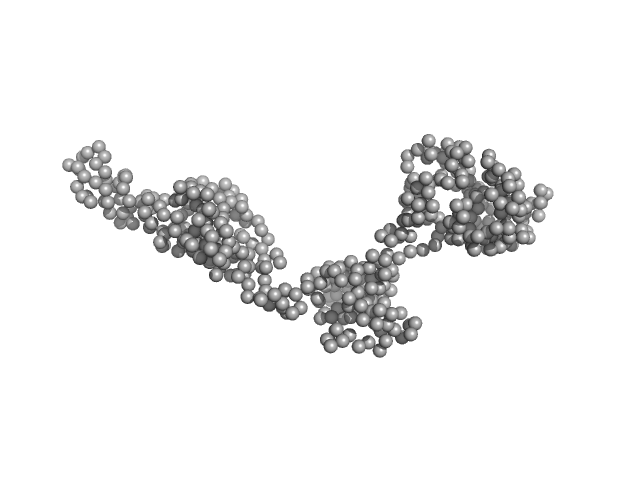
| Sample: |
Uncharacterized protein C1orf159 monomer, 53 kDa Homo sapiens protein
|
| Buffer: |
20mM Hepes 150 NaCl 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Oct 28
|
Probing the Architecture of a Multi-PDZ Domain Protein: Structure of PDZK1 in Solution.
Structure 26(11):1522-1533.e5 (2018)
Hajizadeh NR, Pieprzyk J, Skopintsev P, Flayhan A, Svergun DI, Löw C
|
| RgGuinier |
3.9 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
120 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Transcriptional regulator Lrs14-like protein dimer, 33 kDa Sulfolobus acidocaldarius protein
|
| Buffer: |
300 mM NaCl, 20 mM HEPES, pH 7.5, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 May 5
|
Crystal structure of an Lrs14-like archaeal biofilm regulator from Sulfolobus acidocaldarius.
Acta Crystallogr D Struct Biol 74(Pt 11):1105-1114 (2018)
Vogt MS, Völpel SL, Albers SV, Essen LO, Banerjee A
|
| RgGuinier |
2.9 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
57 |
nm3 |
|
|
|
|
|
|
|
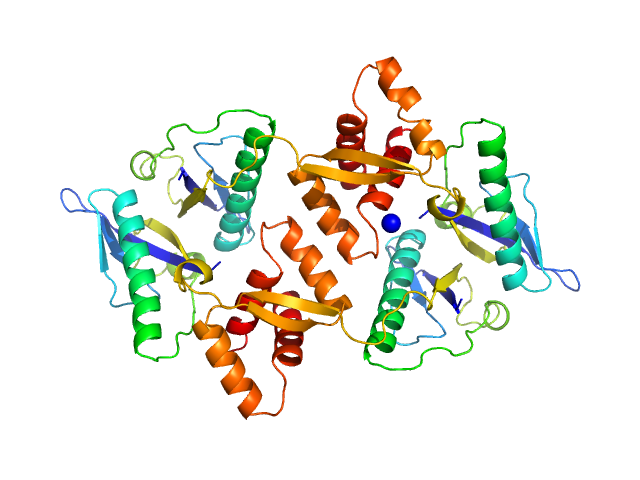
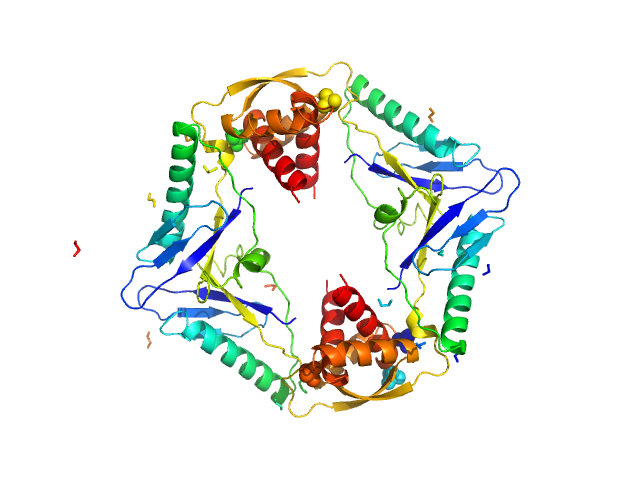
| Sample: |
Type II toxin-antitoxin system HicB family antitoxin tetramer, 63 kDa Burkholderia pseudomallei protein
|
| Buffer: |
25 mM Tris, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2016 Jul 28
|
The molecular basis of protein toxin HicA-dependent binding of the protein antitoxin HicB to DNA.
J Biol Chem (2018)
Winter AJ, Williams C, Isupov MN, Crocker H, Gromova M, Marsh P, Wilkinson OJ, Dillingham MS, Harmer NJ, Titball RW, Crump MP
|
| RgGuinier |
3.0 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
112 |
nm3 |
|
|
|
|
|
|
|
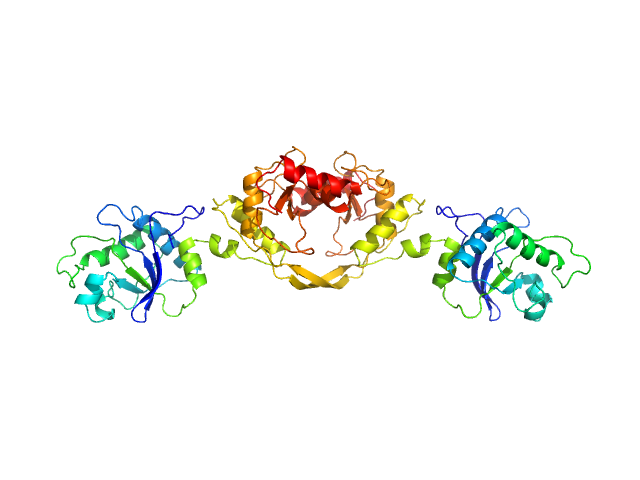
| Sample: |
Type II toxin-antitoxin system HicB family antitoxin tetramer, 63 kDa Burkholderia pseudomallei protein
Addiction module toxin, HicA monomer, 7 kDa Burkholderia pseudomallei protein
|
| Buffer: |
25 mM Tris, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2016 Jul 27
|
The molecular basis of protein toxin HicA-dependent binding of the protein antitoxin HicB to DNA.
J Biol Chem (2018)
Winter AJ, Williams C, Isupov MN, Crocker H, Gromova M, Marsh P, Wilkinson OJ, Dillingham MS, Harmer NJ, Titball RW, Crump MP
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
110 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
5-methylcytosine-specific restriction enzyme A dimer, 65 kDa Escherichia coli protein
|
| Buffer: |
20 mM Tris–HCl pH 7.5, 200 mM KCl, 0.1 mM EDTA, 0.01% (w/v) sodium azide and 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 May 24
|
Activity and structure of EcoKMcrA.
Nucleic Acids Res 46(18):9829-9841 (2018)
Czapinska H, Kowalska M, Zagorskaite E, Manakova E, Slyvka A, Xu SY, Siksnys V, Sasnauskas G, Bochtler M
|
| RgGuinier |
3.7 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
95 |
nm3 |
|
|
|
|
|
|
|
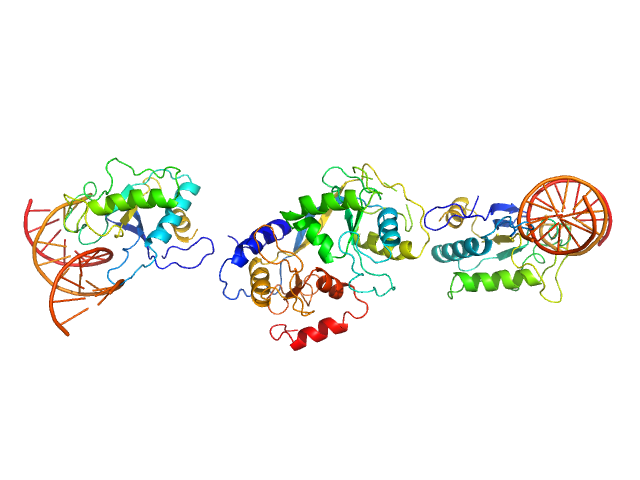
| Sample: |
Cognate hemimethylated 12-bp oligoduplex dimer, 15 kDa DNA
5-methylcytosine-specific restriction enzyme A dimer, 65 kDa Escherichia coli protein
|
| Buffer: |
20 mM Tris–HCl pH 7.5, 200 mM KCl, 0.1 mM EDTA, 0.01% (w/v) sodium azide and 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 May 24
|
Activity and structure of EcoKMcrA.
Nucleic Acids Res 46(18):9829-9841 (2018)
Czapinska H, Kowalska M, Zagorskaite E, Manakova E, Slyvka A, Xu SY, Siksnys V, Sasnauskas G, Bochtler M
|
| RgGuinier |
3.9 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
69 |
nm3 |
|
|
|
|
|
|
|
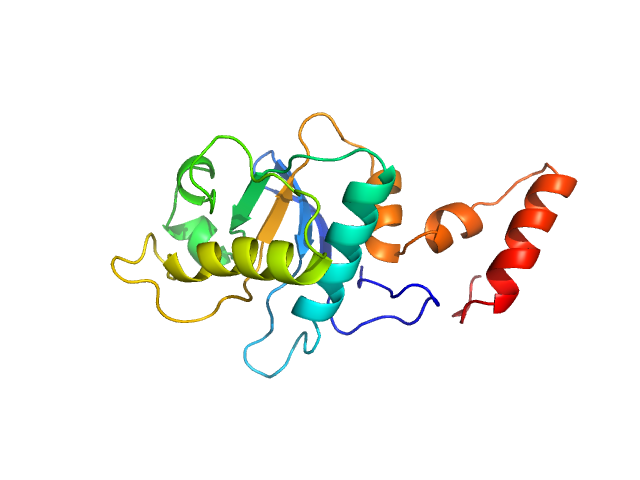
| Sample: |
5-methylcytosine-specific restriction enzyme A (N-terminal domain) monomer, 21 kDa Escherichia coli protein
|
| Buffer: |
20 mM Tris–HCl pH 7.5, 200 mM KCl, 0.1 mM EDTA, 0.01% (w/v) sodium azide, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 May 24
|
Activity and structure of EcoKMcrA.
Nucleic Acids Res 46(18):9829-9841 (2018)
Czapinska H, Kowalska M, Zagorskaite E, Manakova E, Slyvka A, Xu SY, Siksnys V, Sasnauskas G, Bochtler M
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
29 |
nm3 |
|
|
|
|
|
|
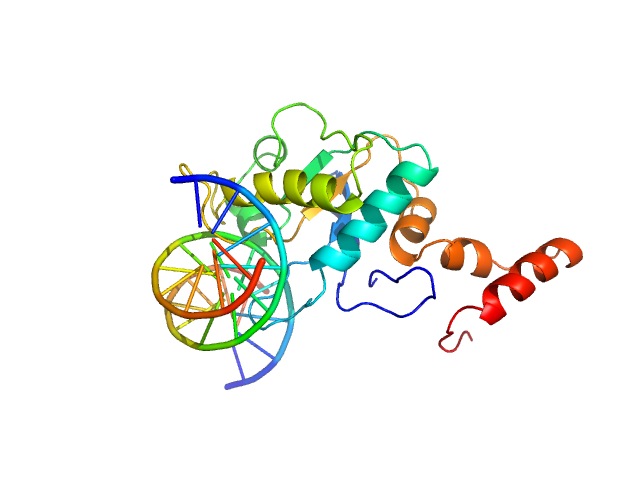
|
| Sample: |
5-methylcytosine-specific restriction enzyme A (N-terminal domain) monomer, 21 kDa Escherichia coli protein
Cognate hemimethylated 12-bp oligoduplex monomer, 7 kDa DNA
|
| Buffer: |
20 mM Tris–HCl pH 7.5, 200 mM KCl, 0.1 mM EDTA, 0.01% (w/v) sodium azide and 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 May 24
|
Activity and structure of EcoKMcrA.
Nucleic Acids Res 46(18):9829-9841 (2018)
Czapinska H, Kowalska M, Zagorskaite E, Manakova E, Slyvka A, Xu SY, Siksnys V, Sasnauskas G, Bochtler M
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
29 |
nm3 |
|
|