|
|
|
|
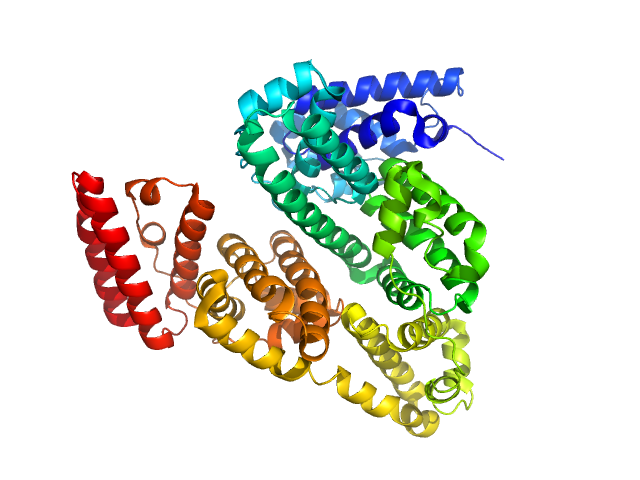
|
| Sample: |
Bovine serum albumin monomer, 66 kDa Bos taurus protein
|
| Buffer: |
PBS, pH: 7.4 |
| Experiment: |
SAXS
data collected at Xenocs BioXolver L with MetalJet, University of Copenhagen, Department of Drug Design and Pharmacology on 2018 Jan 23
|
Size-exclusion chromatography small-angle X-ray scattering of water soluble proteins on a laboratory instrument.
J Appl Crystallogr 51(Pt 6):1623-1632 (2018)
Bucciarelli S, Midtgaard SR, Nors Pedersen M, Skou S, Arleth L, Vestergaard B
|
| RgGuinier |
2.9 |
nm |
| Dmax |
8.6 |
nm |
| VolumePorod |
74 |
nm3 |
|
|
|
|
|
|
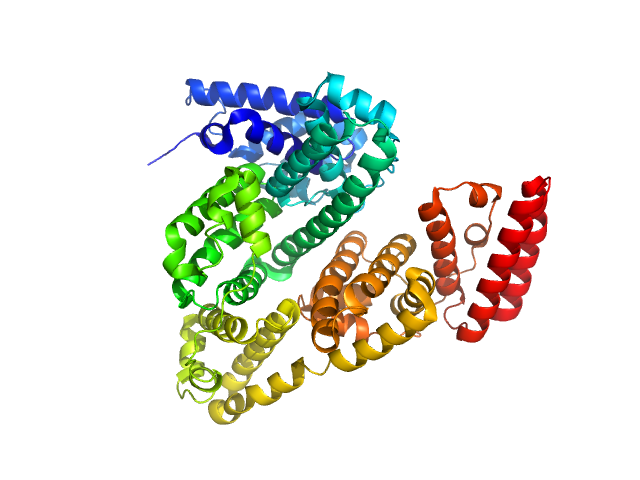
|
| Sample: |
Bovine serum albumin monomer, 66 kDa Bos taurus protein
|
| Buffer: |
PBS, pH: 7.4 |
| Experiment: |
SAXS
data collected at Xenocs BioXolver L with MetalJet, University of Copenhagen, Department of Drug Design and Pharmacology on 2018 Jan 25
|
Size-exclusion chromatography small-angle X-ray scattering of water soluble proteins on a laboratory instrument.
J Appl Crystallogr 51(Pt 6):1623-1632 (2018)
Bucciarelli S, Midtgaard SR, Nors Pedersen M, Skou S, Arleth L, Vestergaard B
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.6 |
nm |
| VolumePorod |
78 |
nm3 |
|
|
|
|
|
|
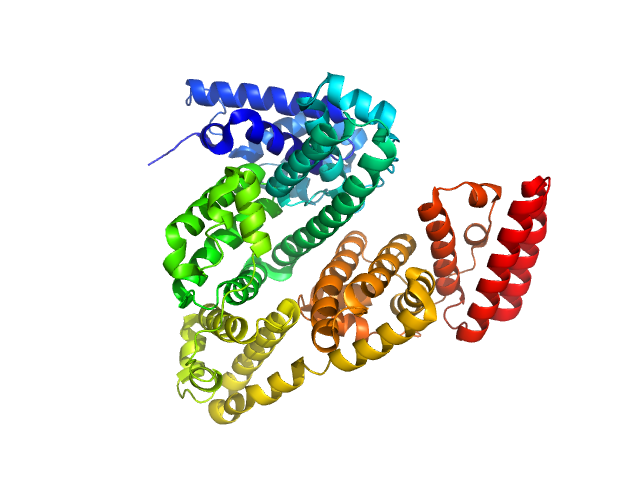
|
| Sample: |
Bovine serum albumin monomer, 66 kDa Bos taurus protein
|
| Buffer: |
50 mM HEPES, pH: 7.5 |
| Experiment: |
SAXS
data collected at Xenocs BioXolver L with MetalJet, University of Copenhagen, Department of Drug Design and Pharmacology on 2018 May 4
|
Size-exclusion chromatography small-angle X-ray scattering of water soluble proteins on a laboratory instrument.
J Appl Crystallogr 51(Pt 6):1623-1632 (2018)
Bucciarelli S, Midtgaard SR, Nors Pedersen M, Skou S, Arleth L, Vestergaard B
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.4 |
nm |
| VolumePorod |
99 |
nm3 |
|
|
|
|
|
|
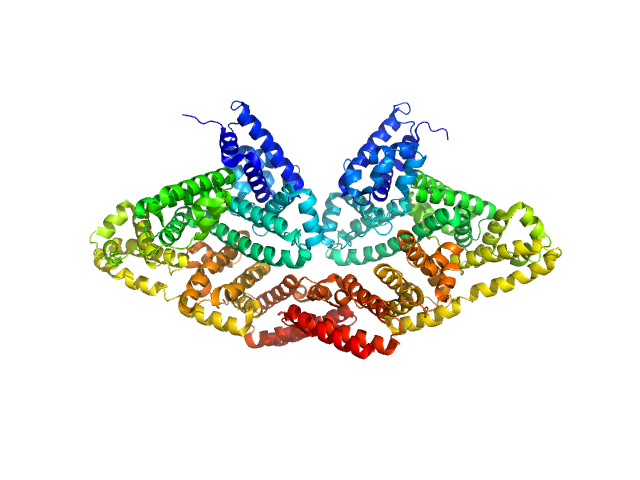
|
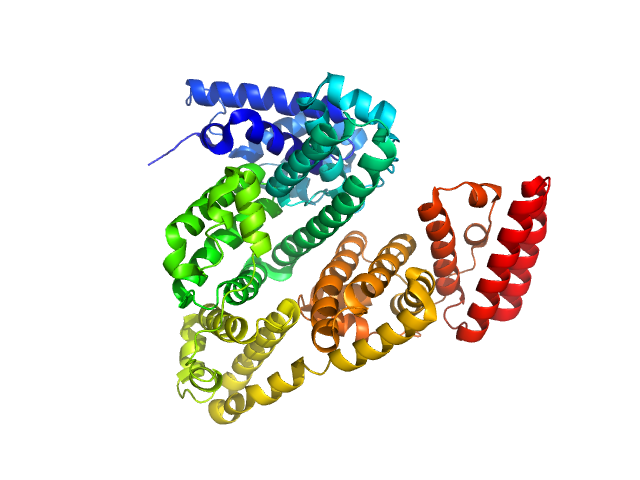
| Sample: |
Bovine serum albumin dimer, 133 kDa Bos taurus protein
|
| Buffer: |
PBS, pH: 7.4 |
| Experiment: |
SAXS
data collected at Xenocs BioXolver L with MetalJet, University of Copenhagen, Department of Drug Design and Pharmacology on 2018 Jan 23
|
Size-exclusion chromatography small-angle X-ray scattering of water soluble proteins on a laboratory instrument.
J Appl Crystallogr 51(Pt 6):1623-1632 (2018)
Bucciarelli S, Midtgaard SR, Nors Pedersen M, Skou S, Arleth L, Vestergaard B
|
| RgGuinier |
4.1 |
nm |
| Dmax |
13.8 |
nm |
| VolumePorod |
200 |
nm3 |
|
|
|
|
|
|
|
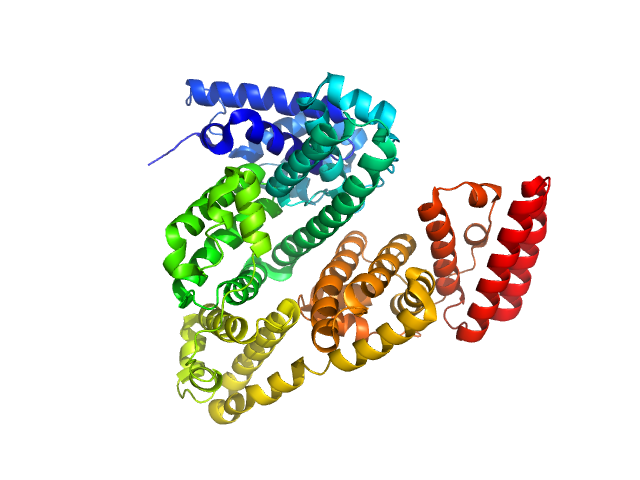
| Sample: |
Bovine serum albumin monomer, 66 kDa Bos taurus protein
|
| Buffer: |
PBS + 1 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Jun 14
|
Size-exclusion chromatography small-angle X-ray scattering of water soluble proteins on a laboratory instrument.
J Appl Crystallogr 51(Pt 6):1623-1632 (2018)
Bucciarelli S, Midtgaard SR, Nors Pedersen M, Skou S, Arleth L, Vestergaard B
|
| RgGuinier |
2.7 |
nm |
| Dmax |
8.7 |
nm |
| VolumePorod |
84 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Bovine serum albumin monomer, 66 kDa Bos taurus protein
|
| Buffer: |
50 mM HEPES, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Nov 26
|
Size-exclusion chromatography small-angle X-ray scattering of water soluble proteins on a laboratory instrument.
J Appl Crystallogr 51(Pt 6):1623-1632 (2018)
Bucciarelli S, Midtgaard SR, Nors Pedersen M, Skou S, Arleth L, Vestergaard B
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
85 |
nm3 |
|
|
|
|
|
|
|
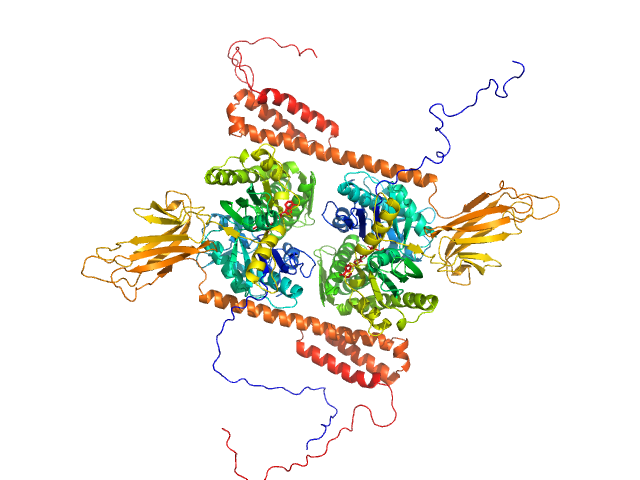
| Sample: |
Heat shock 70 kDa protein 1 dimer, 147 kDa Homo sapiens protein
|
| Buffer: |
50mM HEPES, 150mM KCH3COO, 2mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, CEITEC on 2018 Feb 20
|
Human stress-inducible Hsp70 has a high propensity to form ATP-dependent antiparallel dimers that are differentially regulated by co-chaperone binding.
Mol Cell Proteomics (2018)
Trcka F, Durech M, Vankova P, Chmelik J, Martinkova V, Hausner J, Kadek A, Marcoux J, Klumpler T, Vojtesek B, Muller P, Man P
|
| RgGuinier |
4.0 |
nm |
| Dmax |
11.6 |
nm |
| VolumePorod |
248 |
nm3 |
|
|
|
|
|
|
|
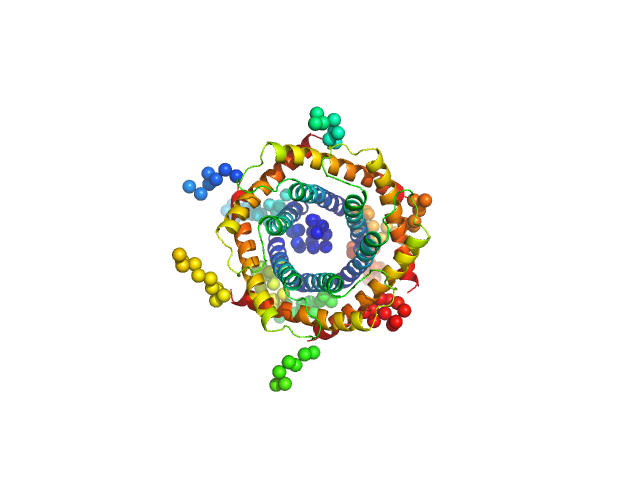
| Sample: |
Mitochondrial import inner membrane translocase subunit TIM9 trimer, 31 kDa Saccharomyces cerevisiae protein
Mitochondrial import inner membrane translocase subunit TIM10 trimer, 31 kDa Saccharomyces cerevisiae protein
|
| Buffer: |
50mM Tris, 150mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Aug 27
|
Structural Basis of Membrane Protein Chaperoning through the Mitochondrial Intermembrane Space.
Cell 175(5):1365-1379.e25 (2018)
Weinhäupl K, Lindau C, Hessel A, Wang Y, Schütze C, Jores T, Melchionda L, Schönfisch B, Kalbacher H, Bersch B, Rapaport D, Brennich M, Lindorff-Larsen K, Wiedemann N, Schanda P
|
| RgGuinier |
2.7 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
103 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Mitochondrial import inner membrane translocase subunit TIM9 hexamer, 61 kDa Saccharomyces cerevisiae protein
Mitochondrial import inner membrane translocase subunit TIM10 hexamer, 61 kDa Saccharomyces cerevisiae protein
Mitochondrial GTP/GDP carrier protein 1 monomer, 33 kDa Saccharomyces cerevisiae protein
|
| Buffer: |
50mM Tris, 150mM NaCl, imidiazole, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Feb 22
|
Structural Basis of Membrane Protein Chaperoning through the Mitochondrial Intermembrane Space.
Cell 175(5):1365-1379.e25 (2018)
Weinhäupl K, Lindau C, Hessel A, Wang Y, Schütze C, Jores T, Melchionda L, Schönfisch B, Kalbacher H, Bersch B, Rapaport D, Brennich M, Lindorff-Larsen K, Wiedemann N, Schanda P
|
| RgGuinier |
4.5 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
272 |
nm3 |
|
|
|
|
|
|
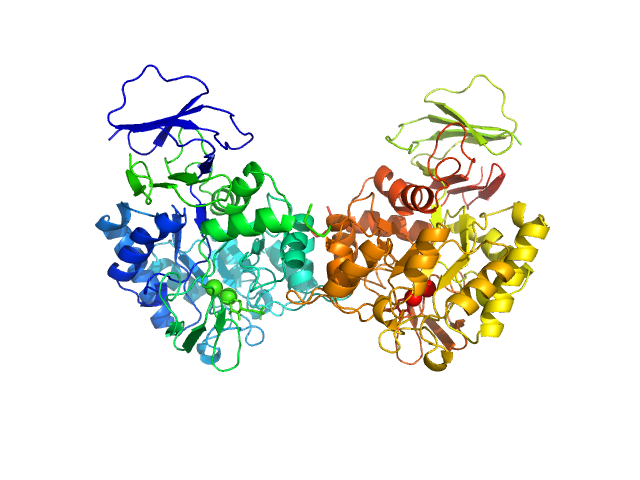
|
| Sample: |
N-acetylglucosamine-6-phosphate deacetylase dimer, 86 kDa Staphylococcus aureus protein
|
| Buffer: |
20 mM Tris-HCl 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2016 Apr 26
|
Functional and solution structure studies of amino sugar deacetylase and deaminase enzymes from Staphylococcus aureus.
FEBS Lett (2018)
Davies JS, Coombes D, Horne CR, Pearce FG, Friemann R, North RA, Dobson RCJ
|
| RgGuinier |
3.2 |
nm |
| Dmax |
10.1 |
nm |
| VolumePorod |
107 |
nm3 |
|
|