|
|
|
|
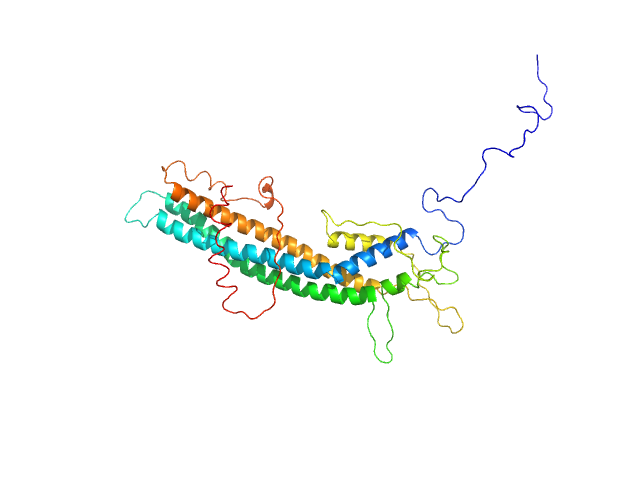
|
| Sample: |
T. brucei spp.-specific protein monomer, 38 kDa Trypanosoma brucei gambiense … protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 3% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2022 Sep 8
|
Beyond the VSG Layer: Exploring the Role of Intrinsic Disorder in the Invariant Surface Glycoproteins of African Trypanosomes.
PLoS Pathog 20(4):e1012186 (2024)
Sülzen H, Volkov AN, Geens R, Zahedifard F, Stijlemans B, Zoltner M, Magez S, Sterckx YG, Zoll S
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
53 |
nm3 |
|
|
|
|
|
|
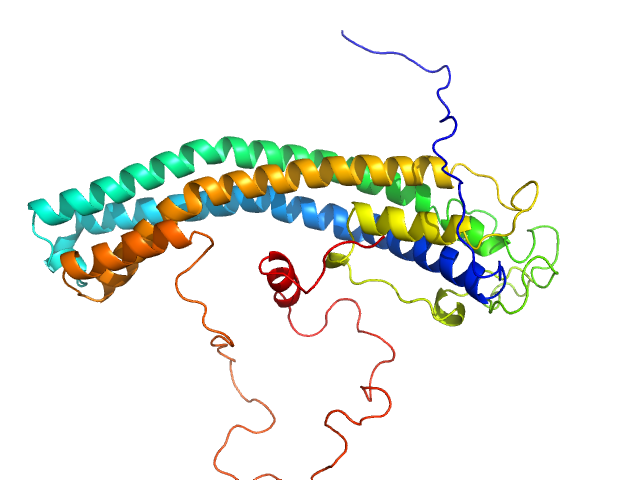
|
| Sample: |
64 kDa invariant surface glycoprotein, putative monomer, 40 kDa Trypanosoma brucei gambiense … protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 3% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2022 Sep 8
|
Beyond the VSG Layer: Exploring the Role of Intrinsic Disorder in the Invariant Surface Glycoproteins of African Trypanosomes.
PLoS Pathog 20(4):e1012186 (2024)
Sülzen H, Volkov AN, Geens R, Zahedifard F, Stijlemans B, Zoltner M, Magez S, Sterckx YG, Zoll S
|
| RgGuinier |
3.1 |
nm |
| Dmax |
11.2 |
nm |
| VolumePorod |
56 |
nm3 |
|
|
|
|
|
|
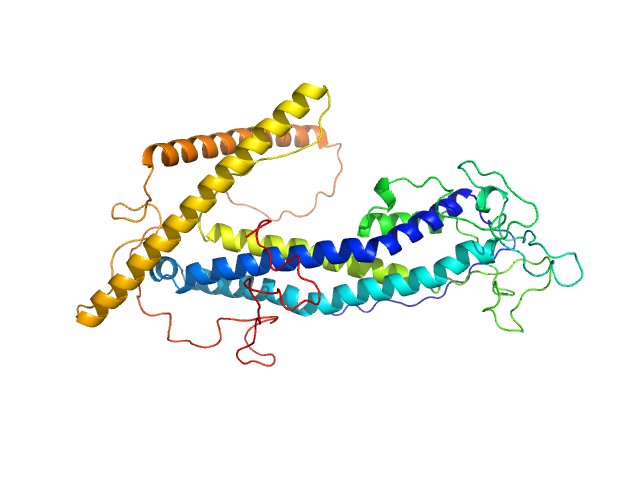
|
| Sample: |
ISG75 monomer, 50 kDa Trypanosoma brucei gambiense protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 3% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2020 Nov 25
|
Beyond the VSG Layer: Exploring the Role of Intrinsic Disorder in the Invariant Surface Glycoproteins of African Trypanosomes.
PLoS Pathog 20(4):e1012186 (2024)
Sülzen H, Volkov AN, Geens R, Zahedifard F, Stijlemans B, Zoltner M, Magez S, Sterckx YG, Zoll S
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.1 |
nm |
| VolumePorod |
85 |
nm3 |
|
|
|
|
|
|
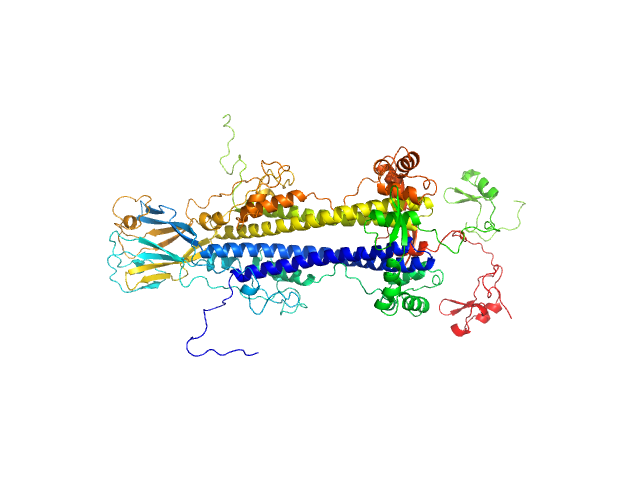
|
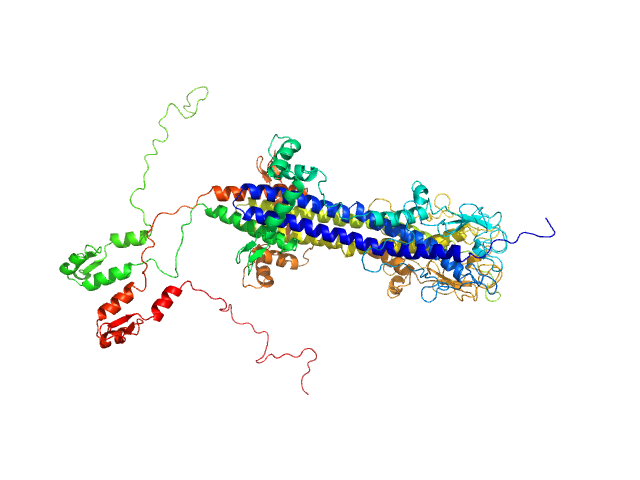
| Sample: |
Variable surface glycoprotein LiTat 1.5 dimer, 98 kDa Trypanosoma brucei gambiense protein
|
| Buffer: |
50 mM Tris-HCl, 500 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2022 Sep 8
|
Beyond the VSG Layer: Exploring the Role of Intrinsic Disorder in the Invariant Surface Glycoproteins of African Trypanosomes.
PLoS Pathog 20(4):e1012186 (2024)
Sülzen H, Volkov AN, Geens R, Zahedifard F, Stijlemans B, Zoltner M, Magez S, Sterckx YG, Zoll S
|
| RgGuinier |
4.4 |
nm |
| Dmax |
17.2 |
nm |
| VolumePorod |
170 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Variant surface glycoprotein 3.1 dimer, 103 kDa Trypanosoma brucei gambiense protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 3% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2022 Sep 8
|
Beyond the VSG Layer: Exploring the Role of Intrinsic Disorder in the Invariant Surface Glycoproteins of African Trypanosomes.
PLoS Pathog 20(4):e1012186 (2024)
Sülzen H, Volkov AN, Geens R, Zahedifard F, Stijlemans B, Zoltner M, Magez S, Sterckx YG, Zoll S
|
| RgGuinier |
4.7 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
149 |
nm3 |
|
|
|
|
|
|
|
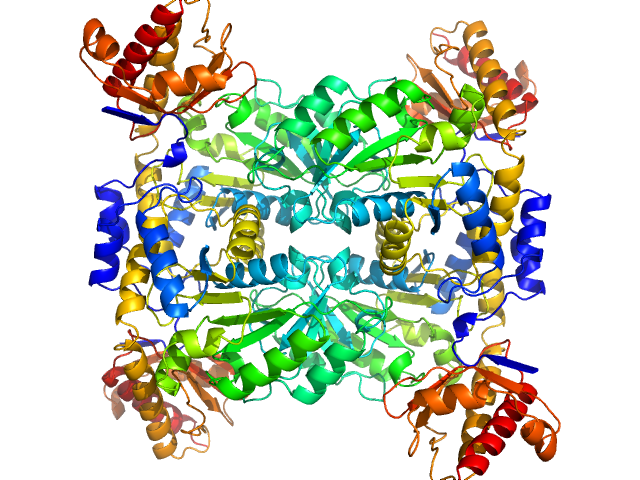
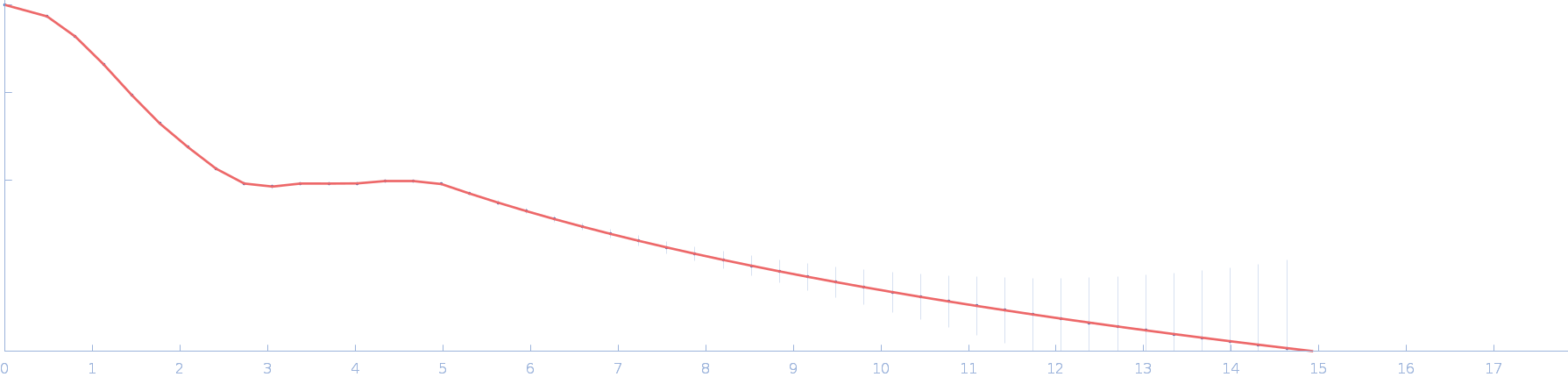
| Sample: |
L-threonine aldolase tetramer, 165 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 1 mM EDTA, 1 mM DTT, pH: 7.8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2023 Feb 14
|
One substrate many enzymes virtual screening uncovers missing genes of carnitine biosynthesis in human and mouse.
Nat Commun 15(1):3199 (2024)
Malatesta M, Fornasier E, Di Salvo ML, Tramonti A, Zangelmi E, Peracchi A, Secchi A, Polverini E, Giachin G, Battistutta R, Contestabile R, Percudani R
|
| RgGuinier |
3.7 |
nm |
| Dmax |
10.6 |
nm |
| VolumePorod |
209 |
nm3 |
|
|
|
|
|
|
|
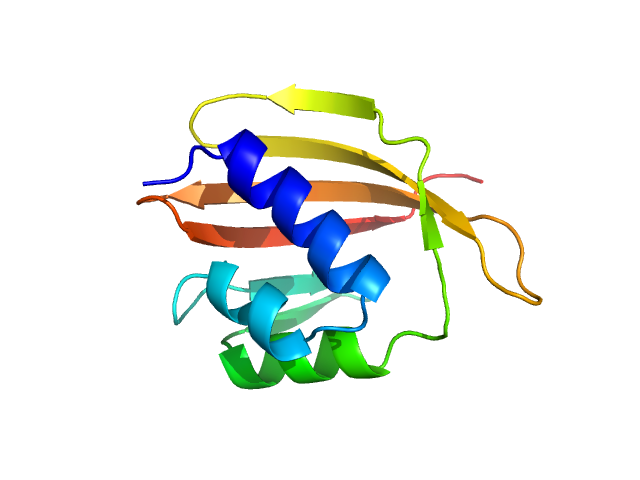
| Sample: |
Ssr1698 protein (H21A) dimer, 22 kDa Synechocystis sp. (strain … protein
|
| Buffer: |
50 mM Hepes, 200 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2022 Dec 7
|
A hemoprotein with a zinc-mirror heme site ties heme availability to carbon metabolism in cyanobacteria.
Nat Commun 15(1):3167 (2024)
Grosjean N, Yee EF, Kumaran D, Chopra K, Abernathy M, Biswas S, Byrnes J, Kreitler DF, Cheng JF, Ghosh A, Almo SC, Iwai M, Niyogi KK, Pakrasi HB, Sarangi R, van Dam H, Yang L, Blaby IK, Blaby-Haas CE
|
| RgGuinier |
2.0 |
nm |
| Dmax |
6.5 |
nm |
| VolumePorod |
28 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
SnoaL-like domain-containing protein monomer, 12 kDa Sphaerobacter thermophilus (strain … protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 3% glycerol, pH: 7.6 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Nov 6
|
Elements of the C-terminal tail of a C-terminal domain homolog of the Orange Carotenoid Protein determining xanthophyll uptake from liposomes.
Biochim Biophys Acta Bioenerg 1865(3):149043 (2024)
Likkei K, Moldenhauer M, Tavraz NN, Egorkin NA, Slonimskiy YB, Maksimov EG, Sluchanko NN, Friedrich T
|
| RgGuinier |
1.5 |
nm |
| Dmax |
5.0 |
nm |
| VolumePorod |
18 |
nm3 |
|
|
|
|
|
|
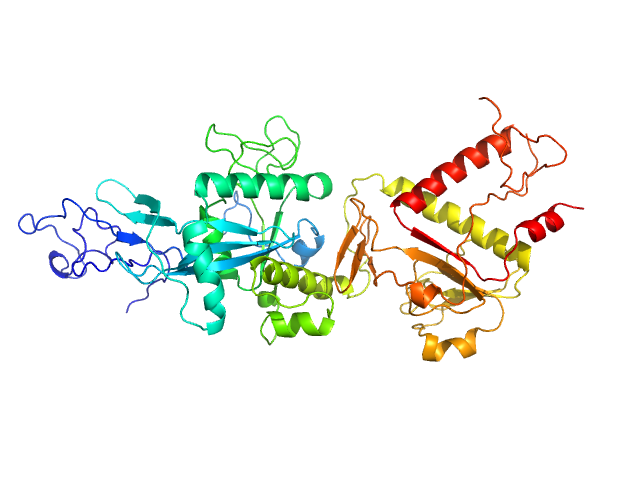
|
| Sample: |
Replicase polyprotein 1ab (non-structural protein 14) monomer, 60 kDa Severe acute respiratory … protein
|
| Buffer: |
50 mM Tris, 150 mM NaCl, 5 mM MgCl2, 2 mM β-mercaptoethanol, pH: 8.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2020 Dec 11
|
Despite the odds: formation of the SARS-CoV-2 methylation complex
Nucleic Acids Research (2024)
Matsuda A, Plewka J, Rawski M, Mourão A, Zajko W, Siebenmorgen T, Kresik L, Lis K, Jones A, Pachota M, Karim A, Hartman K, Nirwal S, Sonani R, Chykunova Y, Minia I, Mak P, Landthaler M, Nowotny M, Dubin G, Sattler M, Suder P, Popowicz G, Pyrć K, Czarna A
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.6 |
nm |
| VolumePorod |
80 |
nm3 |
|
|
|
|
|
|
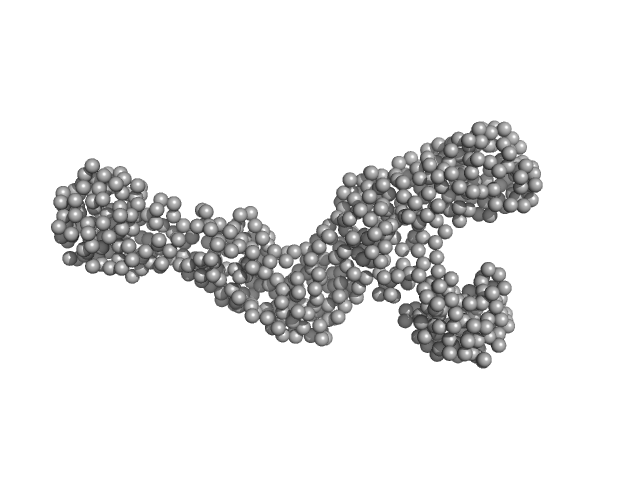
|
| Sample: |
Replicase polyprotein 1ab (non-structural protein 14) monomer, 60 kDa Severe acute respiratory … protein
Replicase polyprotein 1a (non-structural protein 10) monomer, 15 kDa Severe acute respiratory … protein
|
| Buffer: |
50 mM Tris, 150 mM NaCl, 5 mM MgCl2, 2 mM β-mercaptoethanol, pH: 8.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2020 Dec 11
|
Despite the odds: formation of the SARS-CoV-2 methylation complex
Nucleic Acids Research (2024)
Matsuda A, Plewka J, Rawski M, Mourão A, Zajko W, Siebenmorgen T, Kresik L, Lis K, Jones A, Pachota M, Karim A, Hartman K, Nirwal S, Sonani R, Chykunova Y, Minia I, Mak P, Landthaler M, Nowotny M, Dubin G, Sattler M, Suder P, Popowicz G, Pyrć K, Czarna A
|
| RgGuinier |
2.9 |
nm |
| Dmax |
12.2 |
nm |
| VolumePorod |
73 |
nm3 |
|
|