|
|
|
|

|
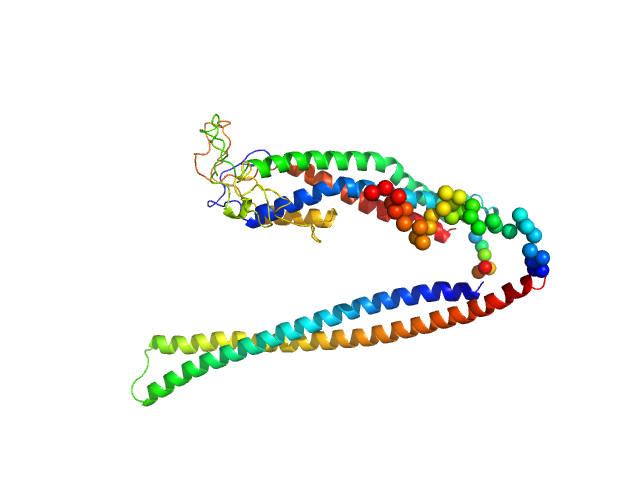
| Sample: |
Replicase polyprotein 1a (non-structural protein 10) monomer, 15 kDa Severe acute respiratory … protein
Replicase polyprotein 1ab (non-structural protein 16) monomer, 33 kDa Severe acute respiratory … protein
|
| Buffer: |
50 mM Tris, 150 mM NaCl, 5 mM MgCl2, 2 mM β-mercaptoethanol, pH: 8.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2020 Dec 11
|
Despite the odds: formation of the SARS-CoV-2 methylation complex
Nucleic Acids Research (2024)
Matsuda A, Plewka J, Rawski M, Mourão A, Zajko W, Siebenmorgen T, Kresik L, Lis K, Jones A, Pachota M, Karim A, Hartman K, Nirwal S, Sonani R, Chykunova Y, Minia I, Mak P, Landthaler M, Nowotny M, Dubin G, Sattler M, Suder P, Popowicz G, Pyrć K, Czarna A
|
| RgGuinier |
2.0 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
28 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Invariant surface glycoprotein (N134A) monomer, 49 kDa Trypanosoma brucei protein
|
| Buffer: |
20 mM Tris-HCl, 75 mM KCl, pH: 7.6 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Aug 18
|
Trypanosoma brucei Invariant Surface Glycoprotein 75 Is an Immunoglobulin Fc Receptor Inhibiting Complement Activation and Antibody-Mediated Cellular Phagocytosis.
J Immunol (2024)
Mikkelsen JH, Stødkilde K, Jensen MP, Hansen AG, Wu Q, Lorentzen J, Graversen JH, Andersen GR, Fenton RA, Etzerodt A, Thiel S, Andersen CBF
|
| RgGuinier |
3.6 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
127 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Alpha domain of Ag43a monomer, 49 kDa Escherichia coli protein
Fragment antigen-binding region Fab10C12 monomer, 47 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2016 Nov 3
|
Inhibition of aggregation and biofilm formation by Uropathogenic Escherichia coli
Andrew Whitten
|
| RgGuinier |
4.3 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
103 |
nm3 |
|
|
|
|
|
|
|
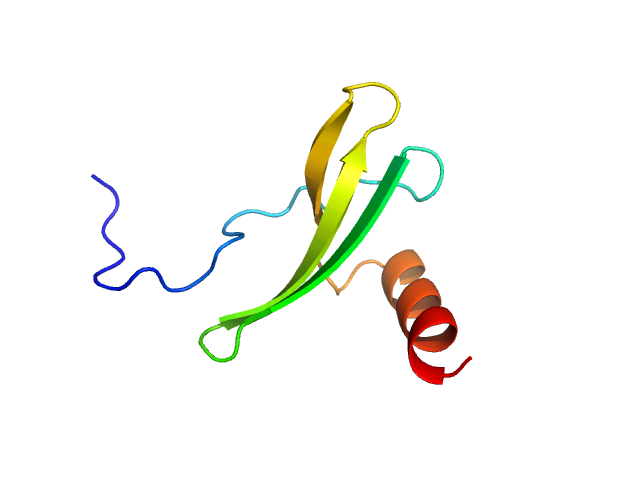
| Sample: |
Stromal cell-derived factor 1 monomer, 9 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 50 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Nov 11
|
The acidic intrinsically disordered region of the inflammatory mediator HMGB1 mediates fuzzy interactions with CXCL12.
Nat Commun 15(1):1201 (2024)
Mantonico MV, De Leo F, Quilici G, Colley LS, De Marchis F, Crippa M, Mezzapelle R, Schulte T, Zucchelli C, Pastorello C, Carmeno C, Caprioglio F, Ricagno S, Giachin G, Ghitti M, Bianchi ME, Musco G
|
| RgGuinier |
1.5 |
nm |
| Dmax |
4.8 |
nm |
| VolumePorod |
19 |
nm3 |
|
|
|
|
|
|

|
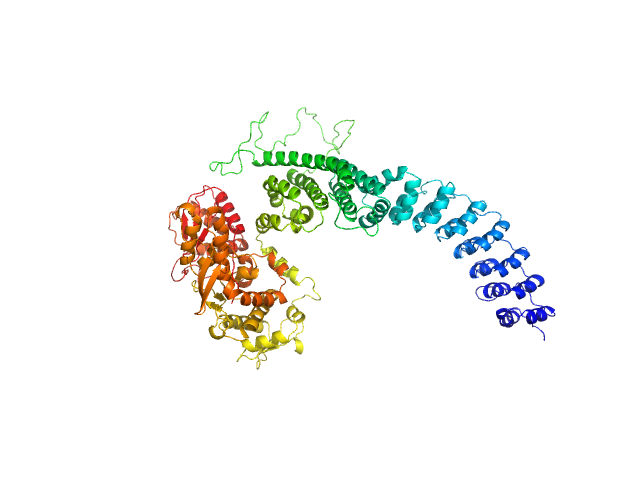
| Sample: |
E3 ubiquitin-protein ligase HACE1 dimer, 205 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 50 mM NaCl, 5 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 Apr 21
|
Structural mechanisms of autoinhibition and substrate recognition by the ubiquitin ligase HACE1
Nature Structural & Molecular Biology (2024)
Düring J, Wolter M, Toplak J, Torres C, Dybkov O, Fokkens T, Bohnsack K, Urlaub H, Steinchen W, Dienemann C, Lorenz S
|
| RgGuinier |
5.2 |
nm |
| Dmax |
16.4 |
nm |
| VolumePorod |
379 |
nm3 |
|
|
|
|
|
|
|
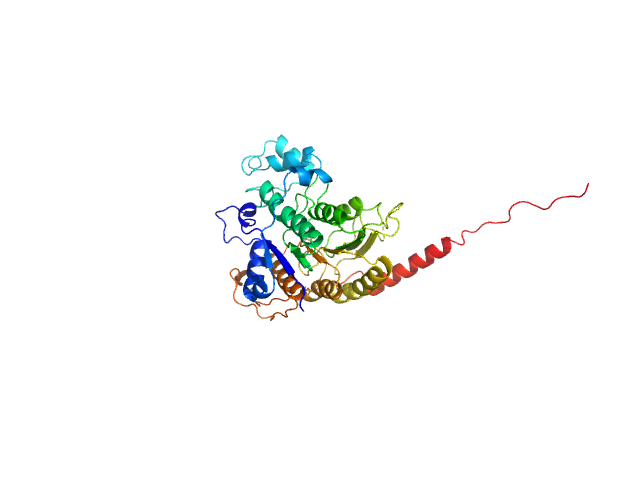
| Sample: |
E3 ubiquitin-protein ligase HACE1 monomer, 100 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 50 mM NaCl, 5 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 Apr 21
|
Structural mechanisms of autoinhibition and substrate recognition by the ubiquitin ligase HACE1
Nature Structural & Molecular Biology (2024)
Düring J, Wolter M, Toplak J, Torres C, Dybkov O, Fokkens T, Bohnsack K, Urlaub H, Steinchen W, Dienemann C, Lorenz S
|
| RgGuinier |
4.6 |
nm |
| Dmax |
17.8 |
nm |
| VolumePorod |
219 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Histone deacetylase 7 monomer, 44 kDa Arabidopsis thaliana protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 1 mM β-mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2023 Jan 24
|
Structure-function analyses reveal Arabidopsis thaliana HDA7 to be an inactive histone deacetylase
Current Research in Structural Biology :100136 (2024)
Saharan K, Baral S, Shaikh N, Vasudevan D
|
| RgGuinier |
3.0 |
nm |
| Dmax |
10.2 |
nm |
| VolumePorod |
85 |
nm3 |
|
|
|
|
|
|
|
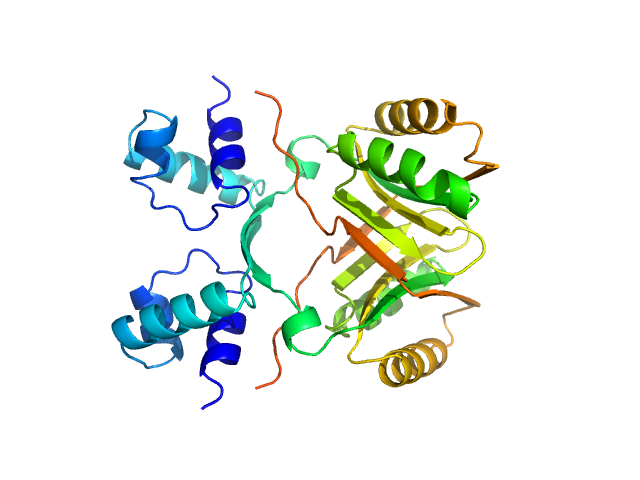
| Sample: |
AsnC family transcriptional regulator dimer, 36 kDa Haloferax mediterranei (strain … protein
|
| Buffer: |
50 mM Tris, 1.5 M NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Nov 12
|
Global Lrp regulator protein from Haloferax mediterranei: Transcriptional analysis and structural characterization.
Int J Biol Macromol :129541 (2024)
Matarredona L, García-Bonete MJ, Guío J, Camacho M, Fillat MF, Esclapez J, Bonete MJ
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.9 |
nm |
| VolumePorod |
70 |
nm3 |
|
|
|
|
|
|
|
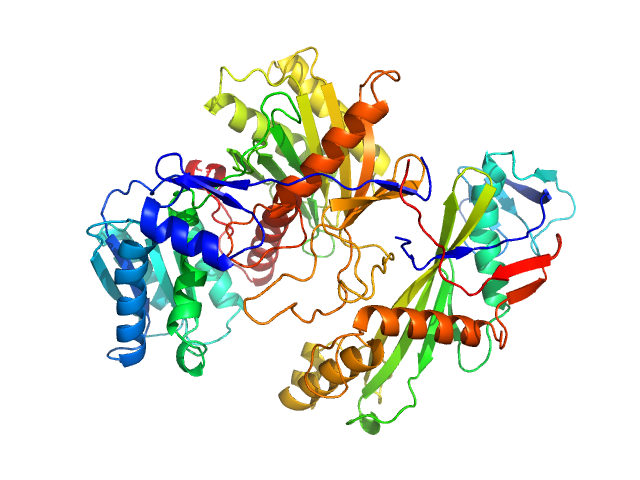
| Sample: |
Piwi protein AF_1318 (Archaeoglobus fulgidus AfAgo protein) monomer, 51 kDa Archaeoglobus fulgidus (strain … protein
Uncharacterized protein (AfAgo-N protein containing N-L1-L2 domains) monomer, 31 kDa Archaeoglobus fulgidus DSM … protein
5'-end phosphorylated DNA oligoduplex, 14 bp (MZ1288) monomer, 9 kDa DNA
|
| Buffer: |
20 mM TrisHCl pH 7.5, 200 mM NaCl, 5 mM MgCl2, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Jul 6
|
The missing part: the Archaeoglobus fulgidus Argonaute forms a functional heterodimer with an N-L1-L2 domain protein.
Nucleic Acids Res (2024)
Manakova E, Golovinas E, Pocevičiūtė R, Sasnauskas G, Silanskas A, Rutkauskas D, Jankunec M, Zagorskaitė E, Jurgelaitis E, Grybauskas A, Venclovas Č, Zaremba M
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
145 |
nm3 |
|
|
|
|
|
|
|
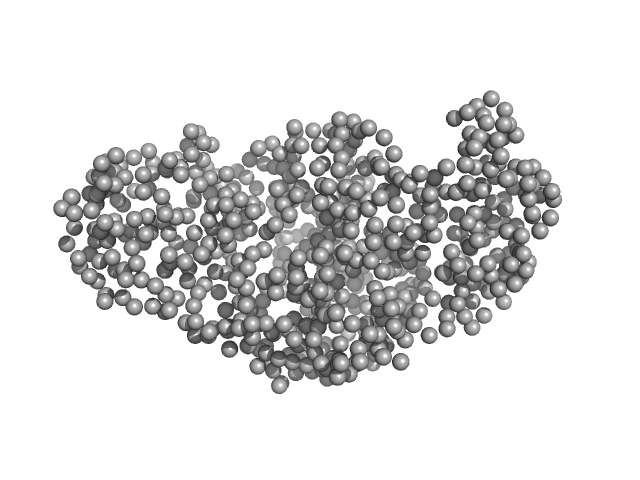
| Sample: |
Single-chain full Archaeoglobus fulgidus Argonaute monomer, 78 kDa Archaeoglobus fulgidus protein
5'-end phosphorylated DNA oligoduplex, 14 bp (MZ1288) monomer, 9 kDa DNA
|
| Buffer: |
20 mM TrisHCl pH7.5, 200 mM NaCl, 5 mM MgCl2, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Jul 6
|
The missing part: the Archaeoglobus fulgidus Argonaute forms a functional heterodimer with an N-L1-L2 domain protein.
Nucleic Acids Res (2024)
Manakova E, Golovinas E, Pocevičiūtė R, Sasnauskas G, Silanskas A, Rutkauskas D, Jankunec M, Zagorskaitė E, Jurgelaitis E, Grybauskas A, Venclovas Č, Zaremba M
|
| RgGuinier |
3.0 |
nm |
| Dmax |
10.3 |
nm |
| VolumePorod |
131 |
nm3 |
|
|