|
|
|
|

|
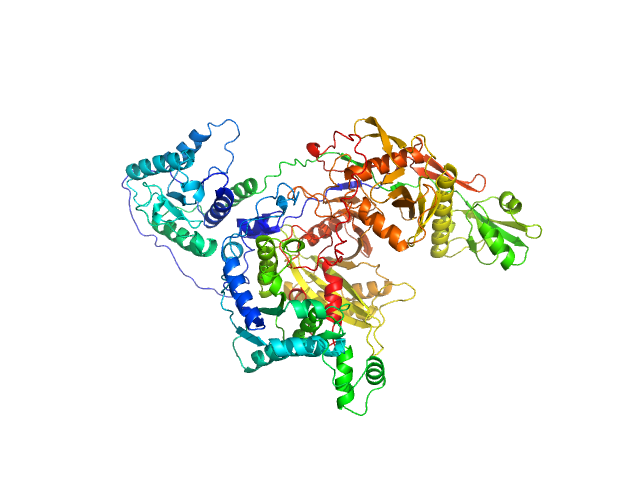
| Sample: |
Codanin-1 monomer, 25 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris HCl, 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpoint 2.0, Institute of Biotechnology, Czech Academy of Sciences/Centre of Molecular Structure on 2022 Mar 5
|
Structural insights into inherited anemia CDA-I: disease-associated mutations disrupt Codanin1-CDIN1 complex
Tomas Brom
|
| RgGuinier |
4.1 |
nm |
| Dmax |
10.7 |
nm |
| VolumePorod |
220 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
TIR domain-containing protein monomer, 57 kDa Rhodopseudomonas palustris (strain … protein
Uncharacterized protein (PIWI) monomer, 56 kDa Rhodopseudomonas palustris (strain … protein
|
| Buffer: |
20 mM Tris-HCl, pH8.0, 200 mM NaCl, 5 mM MgCl2, 2 mM β-mercaptoethanol, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Sep 27
|
Complex of Rhodopseudomonas palustris pAgo with TIR-like effector protein
Elena Manakova
|
| RgGuinier |
3.6 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
192 |
nm3 |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDQZ7_fit2_model1.png)
|
| Sample: |
Ubiquitin carboxyl-terminal hydrolase 14 monomer, 56 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Oct 18
|
Transient interdomain interactions in free USP14 shape its conformational ensemble.
Protein Sci 33(5):e4975 (2024)
Salomonsson J, Wallner B, Sjöstrand L, D'Arcy P, Sunnerhagen M, Ahlner A
|
| RgGuinier |
3.1 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
111 |
nm3 |
|
|
|
|
|
|

|
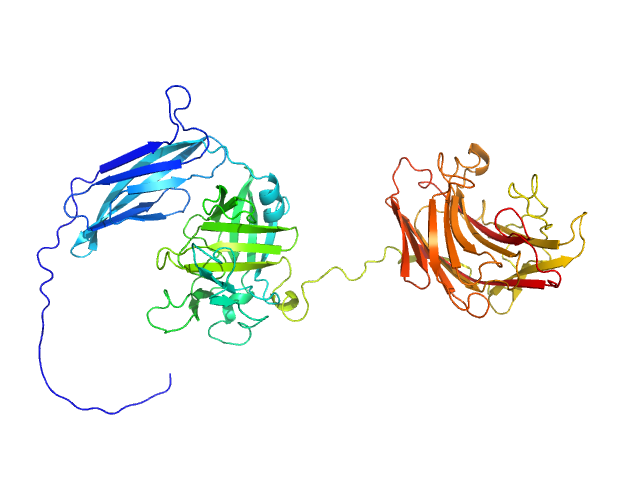
| Sample: |
Ubiquitin carboxyl-terminal hydrolase 14 monomer, 46 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Jul 5
|
Transient interdomain interactions in free USP14 shape its conformational ensemble.
Protein Sci 33(5):e4975 (2024)
Salomonsson J, Wallner B, Sjöstrand L, D'Arcy P, Sunnerhagen M, Ahlner A
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.3 |
nm |
| VolumePorod |
88 |
nm3 |
|
|
|
|
|
|
|
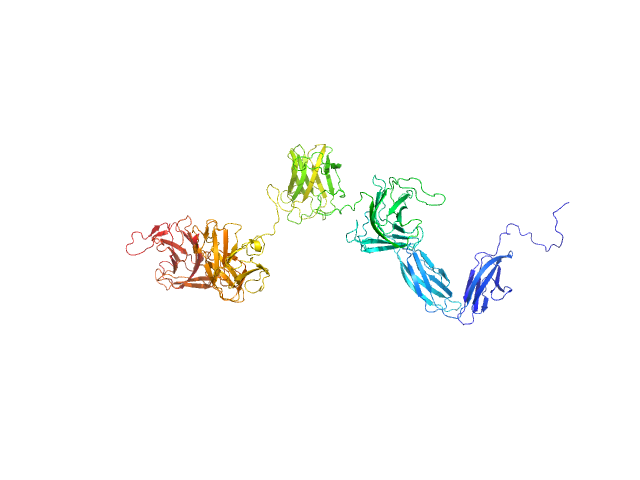
| Sample: |
Glycosyl hydrolase, family 16 monomer, 58 kDa Christiangramia forsetii (strain … protein
|
| Buffer: |
10 mM MOPS, 100 mM NaCl, pH: 7.8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2022 Dec 1
|
Unveiling the role of novel carbohydrate-binding modules in laminarin interaction of multimodular proteins from marine Bacteroidota during phytoplankton blooms.
Environ Microbiol 26(5):e16624 (2024)
Zühlke MK, Ficko-Blean E, Bartosik D, Terrapon N, Jeudy A, Jam M, Wang F, Welsch N, Dürwald A, Martin LT, Larocque R, Jouanneau D, Eisenack T, Thomas F, Trautwein-Schult A, Teeling H, Becher D, Schweder T, Czjzek M
|
| RgGuinier |
3.8 |
nm |
| Dmax |
12.9 |
nm |
| VolumePorod |
76 |
nm3 |
|
|
|
|
|
|
|
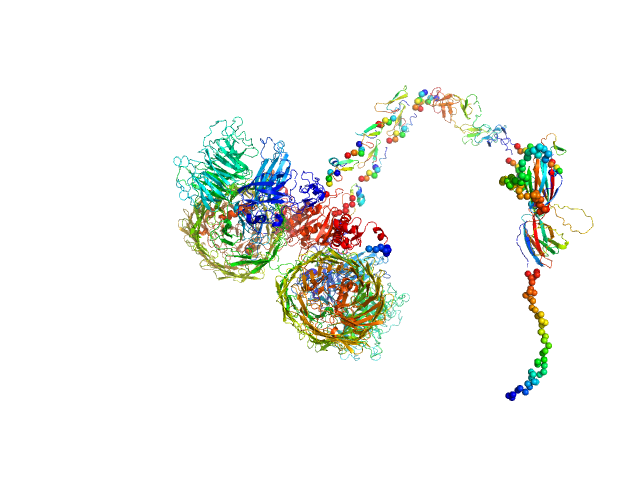
| Sample: |
PKD domain-containing protein monomer, 96 kDa Christiangramia forsetii (strain … protein
|
| Buffer: |
10 mM MOPS, 100 mM NaCl, pH: 7.8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2022 Dec 1
|
Unveiling the role of novel carbohydrate-binding modules in laminarin interaction of multimodular proteins from marine Bacteroidota during phytoplankton blooms.
Environ Microbiol 26(5):e16624 (2024)
Zühlke MK, Ficko-Blean E, Bartosik D, Terrapon N, Jeudy A, Jam M, Wang F, Welsch N, Dürwald A, Martin LT, Larocque R, Jouanneau D, Eisenack T, Thomas F, Trautwein-Schult A, Teeling H, Becher D, Schweder T, Czjzek M
|
| RgGuinier |
5.0 |
nm |
| Dmax |
17.8 |
nm |
| VolumePorod |
138 |
nm3 |
|
|
|
|
|
|
|
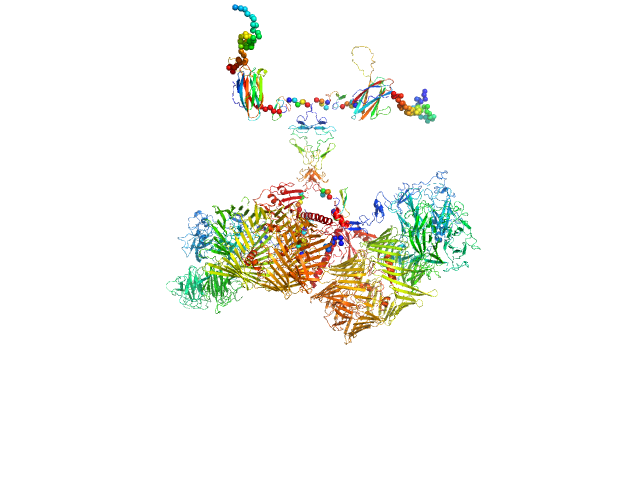
| Sample: |
Isoform A1B1 of Teneurin-3 dimer, 545 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2 mM CaCl2, pH: 7.8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Sep 10
|
Alternative splicing controls teneurin-3 compact dimer formation for neuronal recognition
Nature Communications 15(1) (2024)
Gogou C, Beugelink J, Frias C, Kresik L, Jaroszynska N, Drescher U, Janssen B, Hindges R, Meijer D
|
| RgGuinier |
7.9 |
nm |
| Dmax |
32.0 |
nm |
| VolumePorod |
1129 |
nm3 |
|
|
|
|
|
|
|
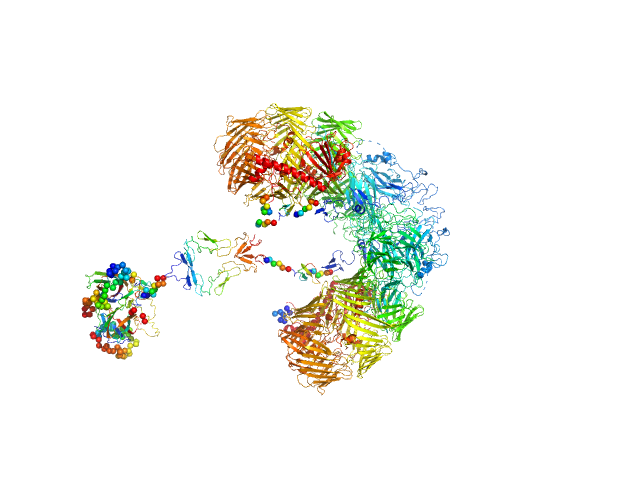
| Sample: |
Isoform A0B0 of Teneurin-3 dimer, 526 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2 mM CaCl2, pH: 7.8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Sep 10
|
Alternative splicing controls teneurin-3 compact dimer formation for neuronal recognition
Nature Communications 15(1) (2024)
Gogou C, Beugelink J, Frias C, Kresik L, Jaroszynska N, Drescher U, Janssen B, Hindges R, Meijer D
|
| RgGuinier |
7.3 |
nm |
| Dmax |
31.0 |
nm |
| VolumePorod |
1057 |
nm3 |
|
|
|
|
|
|
|
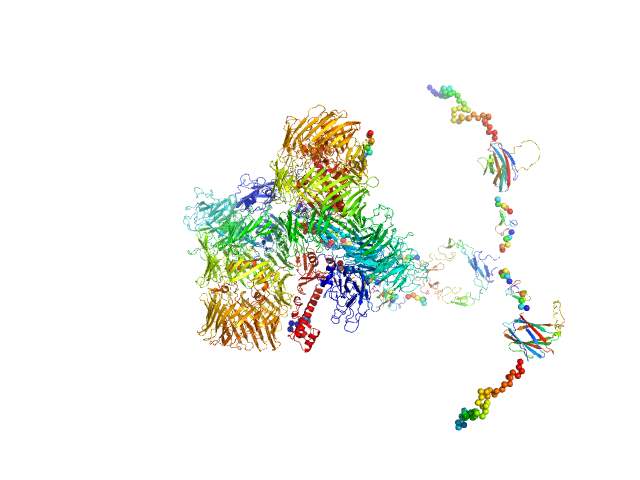
| Sample: |
Isoform A0B1 of Teneurin-3 dimer, 545 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2 mM CaCl2, pH: 7.8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Sep 10
|
Alternative splicing controls teneurin-3 compact dimer formation for neuronal recognition
Nature Communications 15(1) (2024)
Gogou C, Beugelink J, Frias C, Kresik L, Jaroszynska N, Drescher U, Janssen B, Hindges R, Meijer D
|
| RgGuinier |
8.0 |
nm |
| Dmax |
32.0 |
nm |
| VolumePorod |
1053 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Isoform A1B0 of Teneurin-3 dimer, 541 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2 mM CaCl2, pH: 7.8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Sep 10
|
Alternative splicing controls teneurin-3 compact dimer formation for neuronal recognition
Nature Communications 15(1) (2024)
Gogou C, Beugelink J, Frias C, Kresik L, Jaroszynska N, Drescher U, Janssen B, Hindges R, Meijer D
|
| RgGuinier |
7.7 |
nm |
| Dmax |
30.0 |
nm |
| VolumePorod |
1089 |
nm3 |
|
|