|
|
|
|
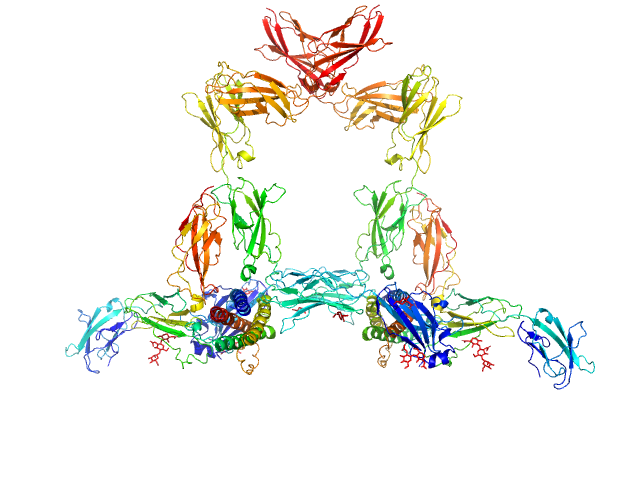
|
| Sample: |
Interleukin-11 dimer, 36 kDa Homo sapiens protein
Interleukin-11 receptor subunit alpha dimer, 64 kDa Homo sapiens protein
Interleukin-6 receptor subunit beta dimer, 134 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 0.2% sodium azide, pH: 8.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Nov 28
|
Structures of the interleukin 11 signalling complex reveal gp130 dynamics and the inhibitory mechanism of a cytokine variant
Nature Communications 14(1) (2023)
Metcalfe R, Hanssen E, Fung K, Aizel K, Kosasih C, Zlatic C, Doughty L, Morton C, Leis A, Parker M, Gooley P, Putoczki T, Griffin M
|
| RgGuinier |
6.2 |
nm |
| Dmax |
20.7 |
nm |
| VolumePorod |
710 |
nm3 |
|
|
|
|
|
|
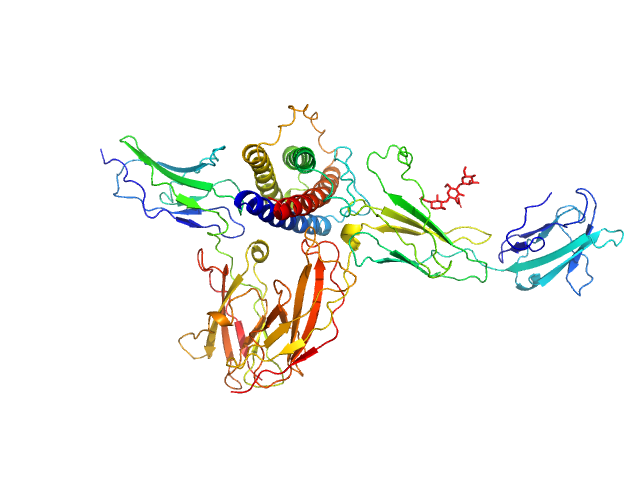
|
| Sample: |
Interleukin-11 receptor subunit alpha monomer, 32 kDa Homo sapiens protein
Interleukin 11 monomer, 18 kDa Homo sapiens protein
Interleukin-6 receptor subunit beta monomer, 23 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 0.2% sodium azide, pH: 8.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Jun 8
|
Structures of the interleukin 11 signalling complex reveal gp130 dynamics and the inhibitory mechanism of a cytokine variant
Nature Communications 14(1) (2023)
Metcalfe R, Hanssen E, Fung K, Aizel K, Kosasih C, Zlatic C, Doughty L, Morton C, Leis A, Parker M, Gooley P, Putoczki T, Griffin M
|
| RgGuinier |
3.6 |
nm |
| Dmax |
12.9 |
nm |
| VolumePorod |
127 |
nm3 |
|
|
|
|
|
|
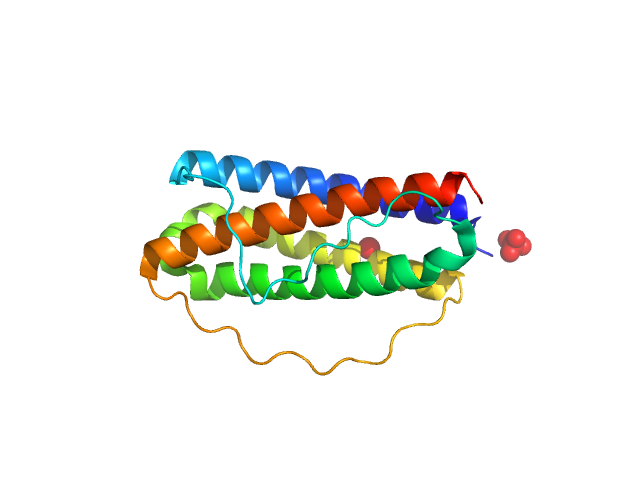
|
| Sample: |
Interleukin-11 (W168A) monomer, 18 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 0.2% sodium azide, pH: 8.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Nov 28
|
Structures of the interleukin 11 signalling complex reveal gp130 dynamics and the inhibitory mechanism of a cytokine variant
Nature Communications 14(1) (2023)
Metcalfe R, Hanssen E, Fung K, Aizel K, Kosasih C, Zlatic C, Doughty L, Morton C, Leis A, Parker M, Gooley P, Putoczki T, Griffin M
|
| RgGuinier |
1.7 |
nm |
| Dmax |
5.2 |
nm |
| VolumePorod |
22 |
nm3 |
|
|
|
|
|
|
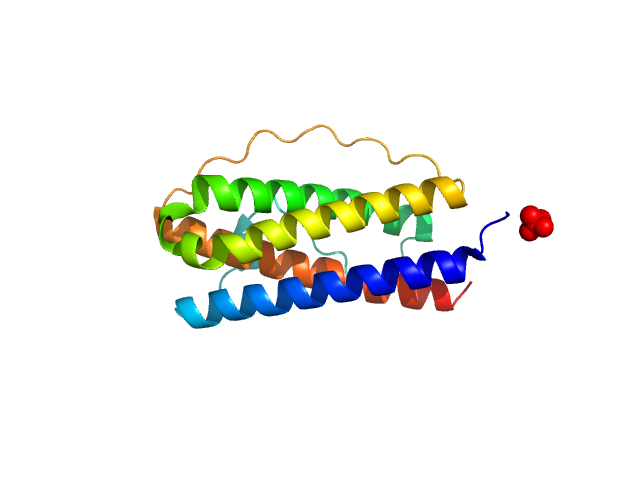
|
| Sample: |
Interleukin 11 Mutein monomer, 18 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 0.2% sodium azide, pH: 8.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Jun 8
|
Structures of the interleukin 11 signalling complex reveal gp130 dynamics and the inhibitory mechanism of a cytokine variant
Nature Communications 14(1) (2023)
Metcalfe R, Hanssen E, Fung K, Aizel K, Kosasih C, Zlatic C, Doughty L, Morton C, Leis A, Parker M, Gooley P, Putoczki T, Griffin M
|
| RgGuinier |
1.8 |
nm |
| Dmax |
5.4 |
nm |
| VolumePorod |
25 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Interleukin-11 receptor subunit alpha monomer, 32 kDa Homo sapiens protein
Interleukin 11 Mutein monomer, 18 kDa Homo sapiens protein
Interleukin-6 receptor subunit beta monomer, 35 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 0.2% sodium azide, pH: 8.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Jun 8
|
Structures of the interleukin 11 signalling complex reveal gp130 dynamics and the inhibitory mechanism of a cytokine variant
Nature Communications 14(1) (2023)
Metcalfe R, Hanssen E, Fung K, Aizel K, Kosasih C, Zlatic C, Doughty L, Morton C, Leis A, Parker M, Gooley P, Putoczki T, Griffin M
|
| RgGuinier |
4.4 |
nm |
| Dmax |
15.6 |
nm |
| VolumePorod |
160 |
nm3 |
|
|
|
|
|
|
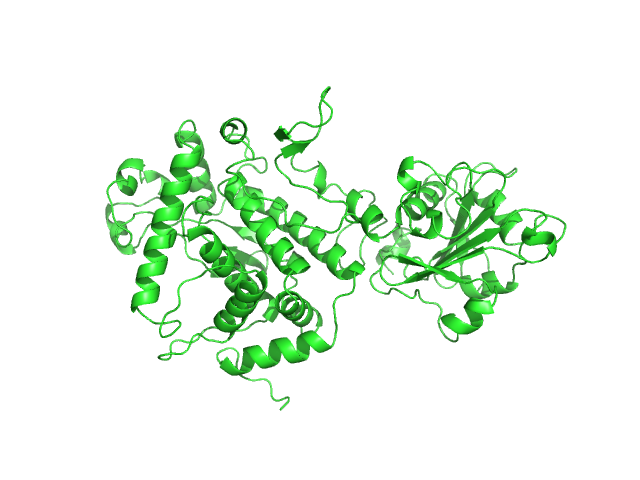
|
| Sample: |
Apoptosis inducing protein monomer, 57 kDa Photobacterium damselae subsp. … protein
|
| Buffer: |
50 mM Hepes, 500 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Apr 13
|
Unconventional structure and mechanisms for membrane interaction and translocation of the NF-κB-targeting toxin AIP56.
Nat Commun 14(1):7431 (2023)
Lisboa J, Pereira C, Pinto RD, Rodrigues IS, Pereira LMG, Pinheiro B, Oliveira P, Pereira PJB, Azevedo JE, Durand D, Benz R, do Vale A, Dos Santos NMS
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
76 |
nm3 |
|
|
|
|
|
|
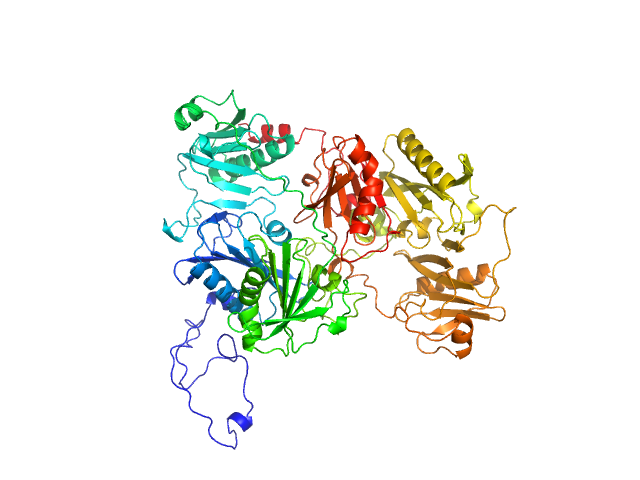
|
| Sample: |
Gelsolin monomer, 85 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 1 mM EDTA, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2020 Feb 24
|
Accurate and Efficient SAXS/SANS Implementation Including Solvation Layer Effects Suitable for Molecular Simulations.
J Chem Theory Comput (2023)
Ballabio F, Paissoni C, Bollati M, de Rosa M, Capelli R, Camilloni C
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.1 |
nm |
| VolumePorod |
128 |
nm3 |
|
|
|
|
|
|
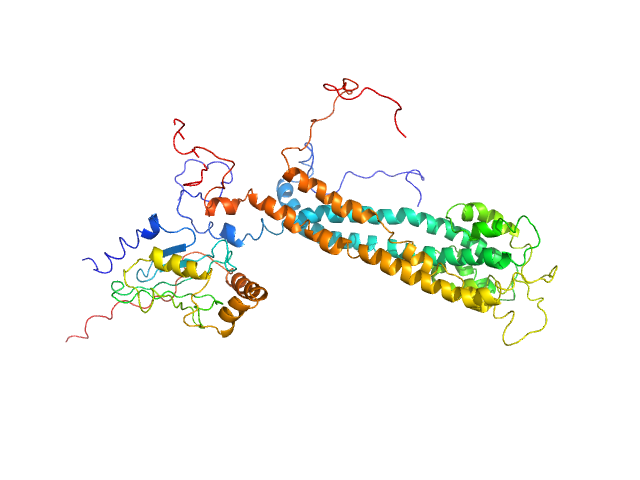
|
| Sample: |
E3 ubiquitin-protein ligase BRE1 dimer, 50 kDa Saccharomyces cerevisiae (strain … protein
Ubiquitin-conjugating enzyme E2 2 monomer, 20 kDa Saccharomyces cerevisiae (strain … protein
|
| Buffer: |
50 mM Tris, 150 mM NaCl, 1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2014 Feb 11
|
Structural basis for the role of C-terminus acidic tail of Saccharomyces cerevisiae ubiquitin-conjugating enzyme (Rad6) in E3 ligase (Bre1) mediated recognition of histones
International Journal of Biological Macromolecules :127717 (2023)
Yadav P, Gupta M, Wazahat R, Islam Z, Tsutakawa S, Kamthan M, Kumar P
|
| RgGuinier |
4.1 |
nm |
| Dmax |
10.4 |
nm |
| VolumePorod |
105 |
nm3 |
|
|
|
|
|
|
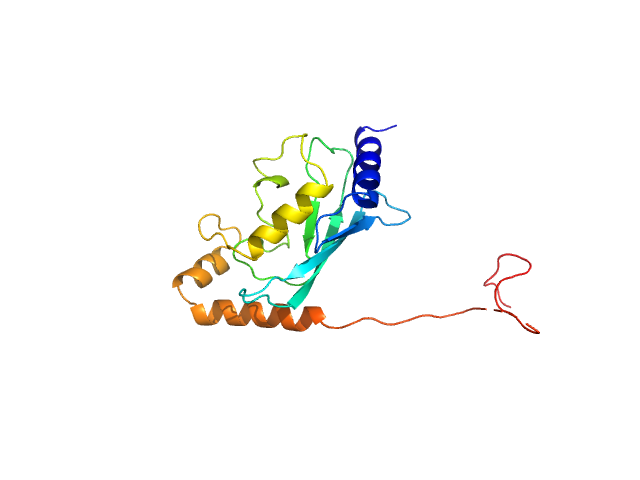
|
| Sample: |
Ubiquitin-conjugating enzyme E2 2 monomer, 20 kDa Saccharomyces cerevisiae (strain … protein
|
| Buffer: |
50 mM Tris, 150 mM NaCl, 1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2014 Feb 11
|
Structural basis for the role of C-terminus acidic tail of Saccharomyces cerevisiae ubiquitin-conjugating enzyme (Rad6) in E3 ligase (Bre1) mediated recognition of histones
International Journal of Biological Macromolecules :127717 (2023)
Yadav P, Gupta M, Wazahat R, Islam Z, Tsutakawa S, Kamthan M, Kumar P
|
| RgGuinier |
2.3 |
nm |
| Dmax |
6.6 |
nm |
| VolumePorod |
31 |
nm3 |
|
|
|
|
|
|
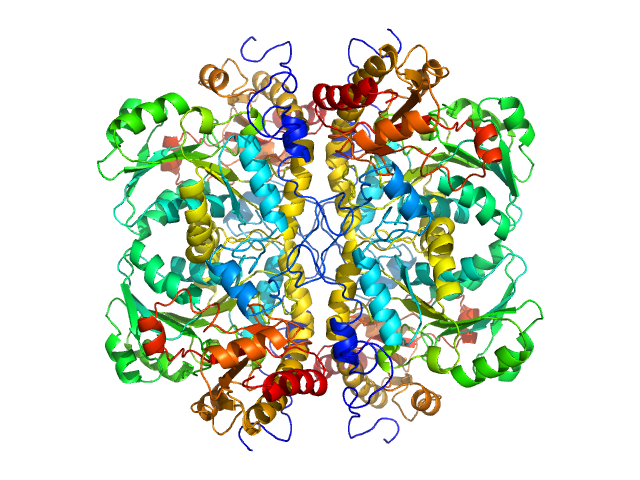
|
| Sample: |
L-methionine gamma-lyase tetramer, 181 kDa Clostridium tetani protein
|
| Buffer: |
PBS-D2O: 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4 (D2O buffer), pH: 7.4 |
| Experiment: |
SANS
data collected at YuMO SANS TOF spectrometer, IBR-2, Frank Laboratory of Neutron Physics, Joint Institute for Nuclear Research on 2019 May 19
|
Methionine gamma lyase fused with S3 domain VGF forms octamers and adheres to tumor cells via binding EGFR
Biochemical and Biophysical Research Communications :149319 (2023)
Bondarev N, Bagaeva D, Bazhenov S, Buben M, Bulushova N, Ryzhykau Y, Okhrimenko I, Zagryadskaya Y, Maslov I, Anisimova N, Sokolova D, Kuklin A, Pokrovsky V, Manukhov I
|
| RgGuinier |
4.0 |
nm |
| Dmax |
14.9 |
nm |
| VolumePorod |
232 |
nm3 |
|
|