|
|
|
|

|
| Sample: |
Precursor microRNA 31 monomer, 23 kDa Homo sapiens RNA
|
| Buffer: |
50 mM potassium phosphate, 50 mM NaCl, 1 mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2023 Mar 19
|
Structure of pre-miR-31 reveals an active role in Dicer-TRBP complex processing.
Proc Natl Acad Sci U S A 120(39):e2300527120 (2023)
Ma S, Kotar A, Hall I, Grote S, Rouskin S, Keane SC
|
| RgGuinier |
2.9 |
nm |
| Dmax |
10.6 |
nm |
| VolumePorod |
31 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Albumin monomer, 66 kDa Bos taurus protein
|
| Buffer: |
10 mM HEPES, 5 mM NaCl, 0.1 mM EDTA, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Jun 8
|
Quantitative size-resolved characterization of mRNA nanoparticles by in-line coupling of asymmetrical-flow field-flow fractionation with small angle X-ray scattering.
Sci Rep 13(1):15764 (2023)
Graewert MA, Wilhelmy C, Bacic T, Schumacher J, Blanchet C, Meier F, Drexel R, Welz R, Kolb B, Bartels K, Nawroth T, Klein T, Svergun D, Langguth P, Haas H
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
104 |
nm3 |
|
|
|
|
|
|
|
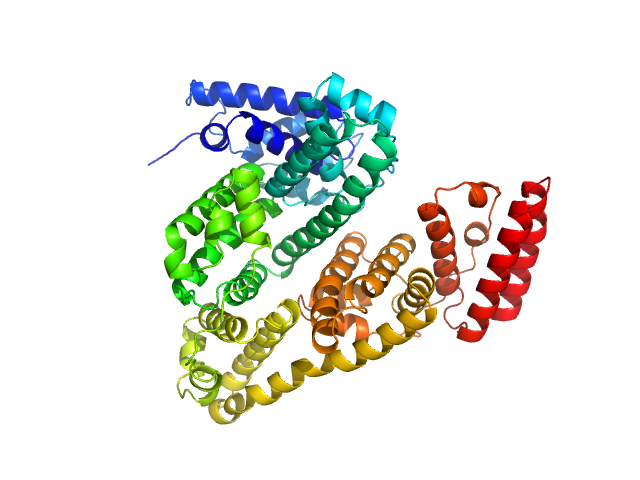
| Sample: |
Endo-D-arabinanase dimer, 107 kDa Microbacterium arabinogalactanolyticum protein
|
| Buffer: |
20 mM Tris-HCl, 200 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2022 Nov 3
|
Identification and characterization of endo-α-, exo-α-, and exo-β-d-arabinofuranosidases degrading lipoarabinomannan and arabinogalactan of mycobacteria
Nature Communications 14(1) (2023)
Shimokawa M, Ishiwata A, Kashima T, Nakashima C, Li J, Fukushima R, Sawai N, Nakamori M, Tanaka Y, Kudo A, Morikami S, Iwanaga N, Akai G, Shimizu N, Arakawa T, Yamada C, Kitahara K, Tanaka K, Ito Y, Fushinobu S, Fujita K
|
| RgGuinier |
3.1 |
nm |
| Dmax |
10.4 |
nm |
| VolumePorod |
131 |
nm3 |
|
|
|
|
|
|
|
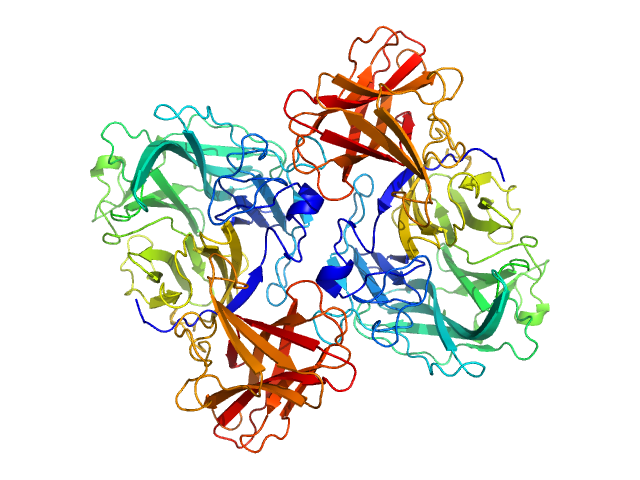
| Sample: |
Glyco_trans_2-like domain-containing protein dimer, 96 kDa Streptomyces sp. M41(2017) protein
|
| Buffer: |
20 mM Tris, pH 7.5, 100 mM NaCl, and 2 mM DTT + 1 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR - Institute of Microbial Technology (IMTech) on 2019 Oct 18
|
Global shape of SvGT, a metal-dependent bacteriocin modifying S/O-HexNActransferase from actinobacteria: c -terminal dimerization modulates the function of this GT.
J Biomol Struct Dyn :1-15 (2023)
Sharma Y, Ahlawat S, Ashish, Rao A
|
| RgGuinier |
4.4 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
151 |
nm3 |
|
|
|
|
|
|
|
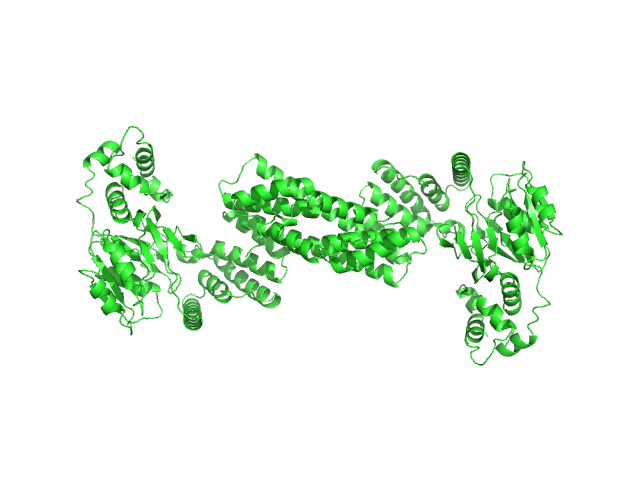
| Sample: |
Glyco_trans_2-like domain-containing protein dimer, 96 kDa Streptomyces sp. M41(2017) protein
|
| Buffer: |
20 mM Tris, pH 7.5, 100 mM NaCl, 2 mM DTT, 1 mM EDTA, 2 mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR - Institute of Microbial Technology (IMTech) on 2019 Oct 18
|
Global shape of SvGT, a metal-dependent bacteriocin modifying S/O-HexNActransferase from actinobacteria: c -terminal dimerization modulates the function of this GT.
J Biomol Struct Dyn :1-15 (2023)
Sharma Y, Ahlawat S, Ashish, Rao A
|
| RgGuinier |
5.8 |
nm |
| Dmax |
17.9 |
nm |
| VolumePorod |
111 |
nm3 |
|
|
|
|
|
|
|
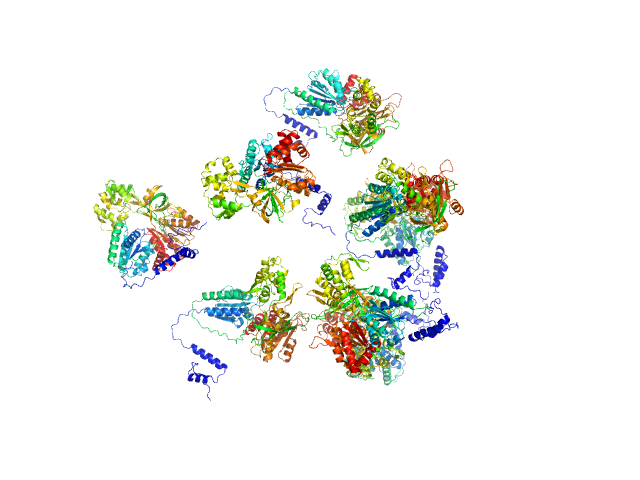
| Sample: |
Sulfite reductase [NADPH] flavoprotein alpha-component octamer, 530 kDa Escherichia coli (strain … protein
|
| Buffer: |
50 mM potassium phosphate, 100 mM NaCl, 1 mM EDTA, pH: 7.8 |
| Experiment: |
SANS
data collected at EQ-SANS, Spallation Neutron Source on 2022 Jun 24
|
Domain crossover in the reductase subunit of NADPH-dependent assimilatory sulfite reductase.
J Struct Biol 215(4):108028 (2023)
Walia N, Murray DT, Garg Y, He H, Weiss KL, Nagy G, Elizabeth Stroupe M
|
| RgGuinier |
8.4 |
nm |
| Dmax |
23.0 |
nm |
| VolumePorod |
767 |
nm3 |
|
|
|
|
|
|
|
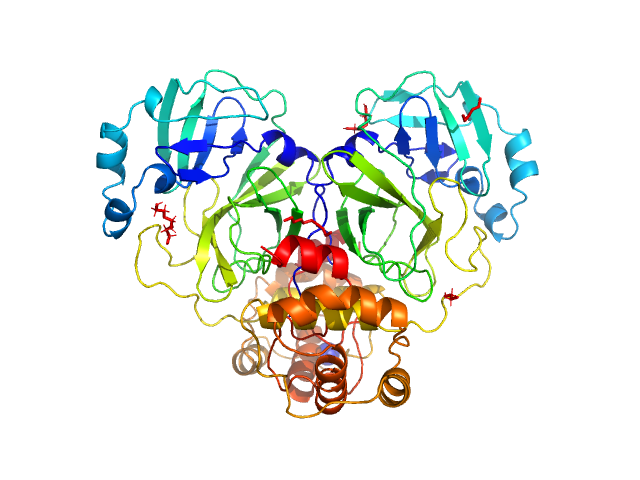
| Sample: |
Replicase polyprotein 1ab, H3426A (3C-like proteinase nsp5 - H163A mutant) dimer, 67 kDa Severe acute respiratory … protein
|
| Buffer: |
25 mM HEPES, 1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2023 Mar 24
|
The H163A mutation unravels an oxidized conformation of the SARS-CoV-2 main protease.
Nat Commun 14(1):5625 (2023)
Tran N, Dasari S, Barwell SAE, McLeod MJ, Kalyaanamoorthy S, Holyoak T, Ganesan A
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
102 |
nm3 |
|
|
|
|
|
|
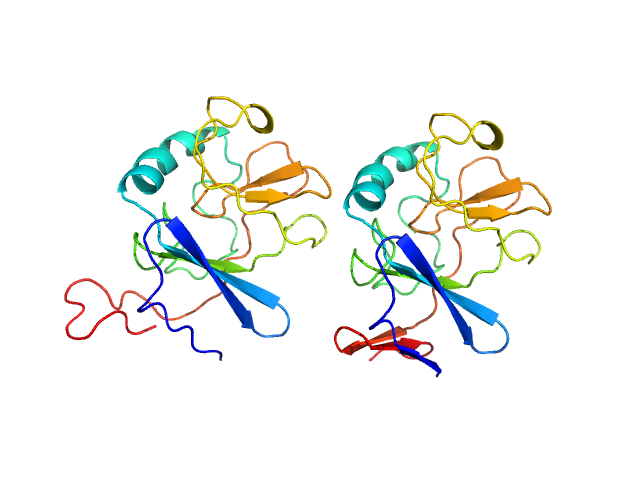
|
| Sample: |
Ubiquinol-cytochrome c reductase iron-sulfur subunit dimer, 33 kDa Thermochromatium tepidum protein
|
| Buffer: |
10 mM Tris-HCl pH 7.6, 150 mM NaCl, 5% glycerol, pH: 7.6 |
| Experiment: |
SAXS
data collected at BL38B1, SPring-8 on 2023 Apr 26
|
Structure of a putative immature form of a Rieske-type iron-sulfur protein in complex with zinc chloride.
Commun Chem 6(1):190 (2023)
Tsutsumi E, Niwa S, Takeda R, Sakamoto N, Okatsu K, Fukai S, Ago H, Nagao S, Sekiguchi H, Takeda K
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.8 |
nm |
| VolumePorod |
38 |
nm3 |
|
|
|
|
|
|
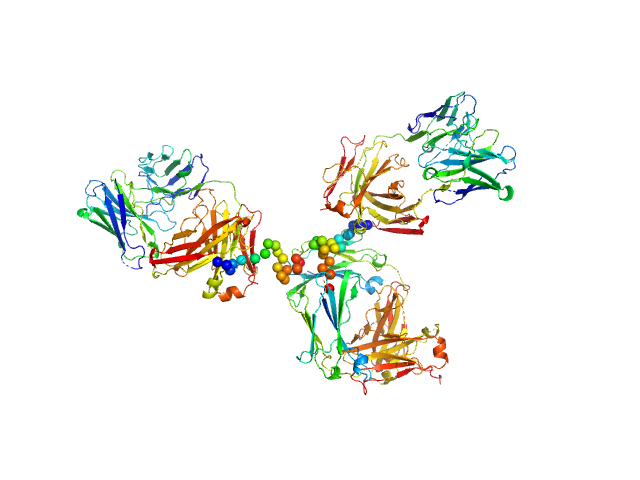
|
| Sample: |
Palivizumab IgG monomer, 145 kDa Commercial (Synagis) protein
|
| Buffer: |
PBS (137 mM NaCl, 2.7 mM KCl, 12 mM HPO4 2−/H2PO4 −), pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Sep 14
|
Respiratory syncytial virus (RSV)-approved monoclonal antibody Palivizumab as ligand for anti-idiotype nanobody-based synthetic cytokine receptors
Journal of Biological Chemistry :105270 (2023)
Ettich J, Wittich C, Moll J, Behnke K, Floss D, Reiners J, Christmann A, Lang P, Smits S, Kolmar H, Scheller J
|
| RgGuinier |
5.0 |
nm |
| Dmax |
16.8 |
nm |
| VolumePorod |
252 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Anti-Idiotypic-Palivizumab-Nanobody1 monomer, 20 kDa Lama glama protein
|
| Buffer: |
PBS (137 mM NaCl, 2.7 mM KCl, 12 mM HPO4 2−/H2PO4 −), pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Sep 14
|
Respiratory syncytial virus (RSV)-approved monoclonal antibody Palivizumab as ligand for anti-idiotype nanobody-based synthetic cytokine receptors
Journal of Biological Chemistry :105270 (2023)
Ettich J, Wittich C, Moll J, Behnke K, Floss D, Reiners J, Christmann A, Lang P, Smits S, Kolmar H, Scheller J
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.8 |
nm |
| VolumePorod |
31 |
nm3 |
|
|