|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDXB4_fit1_model1.png)
|
| Sample: |
Peroxisomal biogenesis factor 8 monomer, 78 kDa Komagataella pastoris protein
Peroxisomal targeting signal receptor monomer, 32 kDa Komagataella pastoris protein
|
| Buffer: |
50 mM HEPES, 150 mM NaCl, 3% v/v glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Aug 17
|
Structure of Pex8 in complex with peroxisomal receptor Pex5 reveals its essential role in peroxisomal cargo translocation
(2025)
Ekal L, Wendscheck D, David Y, Chojnowski G, Jeffries C, Mullapudi E, Schuldiner M, Warscheid B, Zalckvar E, Wilmanns M
|
| RgGuinier |
4.6 |
nm |
| Dmax |
19.0 |
nm |
| VolumePorod |
177 |
nm3 |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDXC4_fit1_model1.png)
|
| Sample: |
Peroxisomal biogenesis factor 8 monomer, 78 kDa Komagataella pastoris protein
Peroxisomal targeting signal receptor monomer, 36 kDa Komagataella pastoris protein
|
| Buffer: |
50 mM HEPES, 150 mM NaCl, 3% v/v glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Aug 17
|
Structure of Pex8 in complex with peroxisomal receptor Pex5 reveals its essential role in peroxisomal cargo translocation
(2025)
Ekal L, Wendscheck D, David Y, Chojnowski G, Jeffries C, Mullapudi E, Schuldiner M, Warscheid B, Zalckvar E, Wilmanns M
|
| RgGuinier |
4.2 |
nm |
| Dmax |
14.5 |
nm |
| VolumePorod |
141 |
nm3 |
|
|
|
|
|
|
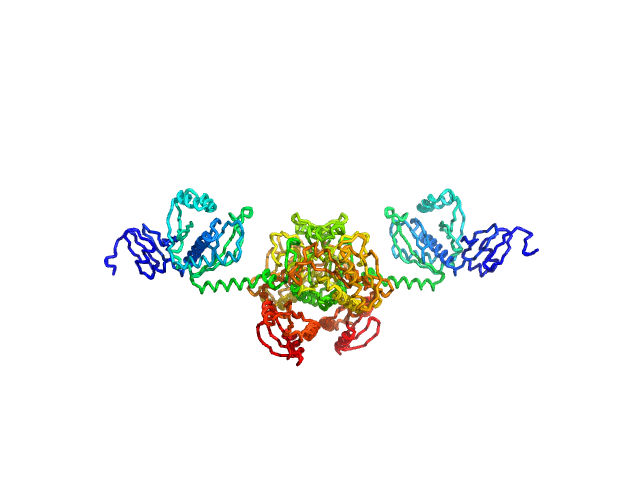
|
| Sample: |
Stenotrophomonas maltophilia threonyl tRNA synthetase dimer, 146 kDa Stenotrophomonas maltophilia (strain … protein
|
| Buffer: |
20 mM HEPES, 0.2 M NaCl, 1.5% v/v glycerol, and 1 mM TCEP, pH: 7 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2024 Nov 7
|
Crystal structure of a complete threonyl tRNA synthetase: toggle clamp model?
Biochemical and Biophysical Research Communications :152677 (2025)
Tran T, Fenwick M, Edwards T, Dranow D, Abendroth J, Shek R, Tillery L, Craig J, Van Voorhis W, Myler P
|
| RgGuinier |
4.2 |
nm |
| Dmax |
16.2 |
nm |
|
|
|
|
|
|
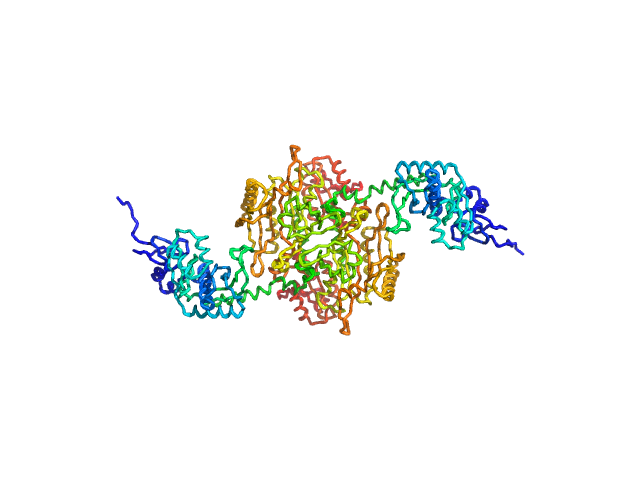
|
| Sample: |
Stenotrophomonas maltophilia threonyl tRNA synthetase dimer, 146 kDa Stenotrophomonas maltophilia (strain … protein
|
| Buffer: |
20 mM HEPES, 0.2 M NaCl, 1.5% v/v glycerol, and 1 mM TCEP, pH: 7 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2024 Nov 7
|
Crystal structure of a complete threonyl tRNA synthetase: toggle clamp model?
Biochemical and Biophysical Research Communications :152677 (2025)
Tran T, Fenwick M, Edwards T, Dranow D, Abendroth J, Shek R, Tillery L, Craig J, Van Voorhis W, Myler P
|
| RgGuinier |
4.2 |
nm |
| Dmax |
16.2 |
nm |
|
|
|
|
|
|
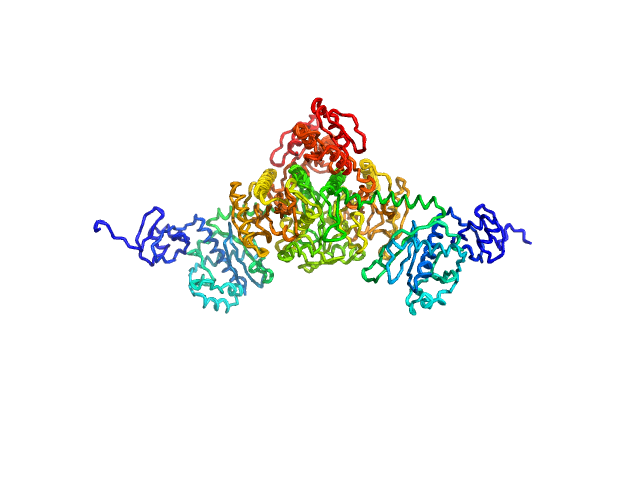
|
| Sample: |
Stenotrophomonas maltophilia threonyl tRNA synthetase dimer, 146 kDa Stenotrophomonas maltophilia (strain … protein
|
| Buffer: |
20 mM HEPES, 0.2 M NaCl, 1.5% v/v glycerol, and 1 mM TCEP, pH: 7 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2024 Nov 7
|
Crystal structure of a complete threonyl tRNA synthetase: toggle clamp model?
Biochemical and Biophysical Research Communications :152677 (2025)
Tran T, Fenwick M, Edwards T, Dranow D, Abendroth J, Shek R, Tillery L, Craig J, Van Voorhis W, Myler P
|
| RgGuinier |
4.2 |
nm |
| Dmax |
16.2 |
nm |
|
|
|
|
|
|
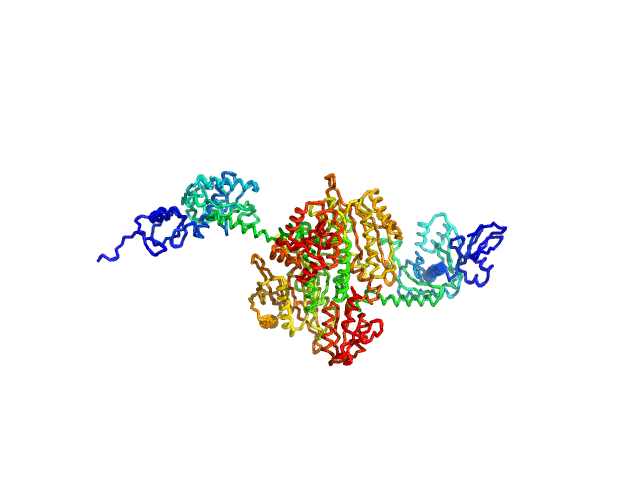
|
| Sample: |
Stenotrophomonas maltophilia threonyl tRNA synthetase dimer, 146 kDa protein
|
| Buffer: |
20 mM HEPES, 0.2 M NaCl, 1.5% v/v glycerol, and 1 mM TCEP, pH: 7 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2024 Nov 7
|
Crystal structure of a complete threonyl tRNA synthetase: toggle clamp model?
Biochemical and Biophysical Research Communications :152677 (2025)
Tran T, Fenwick M, Edwards T, Dranow D, Abendroth J, Shek R, Tillery L, Craig J, Van Voorhis W, Myler P
|
|
|
|
|
|
|
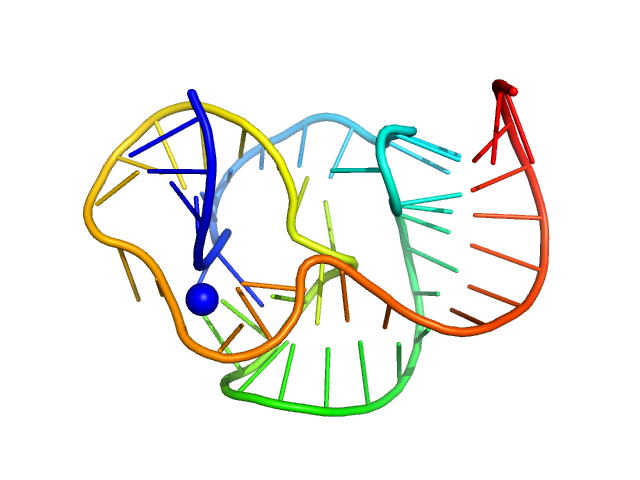
|
| Sample: |
ST9 WT (54 nt RNA) monomer, 17 kDa RNA
|
| Buffer: |
50 mM Tris pH 7.5, 100 mM NaCl, 1 mM DTT, 10 mM MgCl2, 0.0002% w/v NaN3, pH: |
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2024 Mar 23
|
A conserved viral RNA fold enables nuclease resistance across kingdoms of life.
Nucleic Acids Res 53(16) (2025)
Gezelle JG, Korn SM, McDonald JT, Gong Z, Erickson A, Huang CH, Yang F, Cronin M, Kuo YW, Wimberly BT, Steckelberg AL
|
| RgGuinier |
1.9 |
nm |
| Dmax |
5.8 |
nm |
| VolumePorod |
21 |
nm3 |
|
|
|
|
|
|
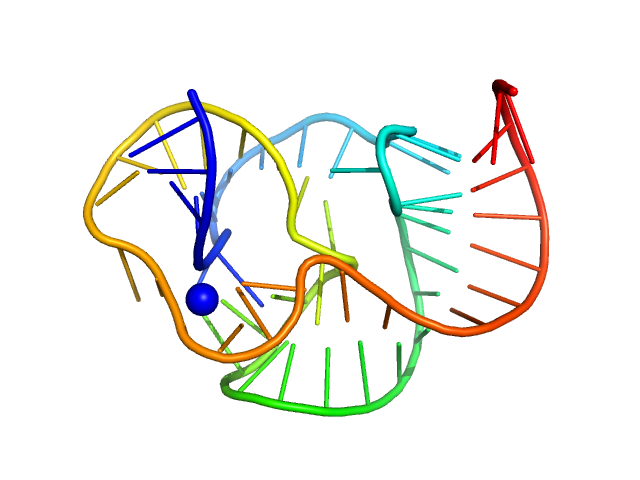
|
| Sample: |
ST9 WT (54 nt RNA) monomer, 17 kDa RNA
|
| Buffer: |
50 mM Tris pH 7.5, 100 mM NaCl, 1 mM DTT, 1 mM MgCl2, pH: |
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2024 Mar 23
|
A conserved viral RNA fold enables nuclease resistance across kingdoms of life.
Nucleic Acids Res 53(16) (2025)
Gezelle JG, Korn SM, McDonald JT, Gong Z, Erickson A, Huang CH, Yang F, Cronin M, Kuo YW, Wimberly BT, Steckelberg AL
|
| RgGuinier |
1.7 |
nm |
| Dmax |
5.8 |
nm |
| VolumePorod |
20 |
nm3 |
|
|
|
|
|
|
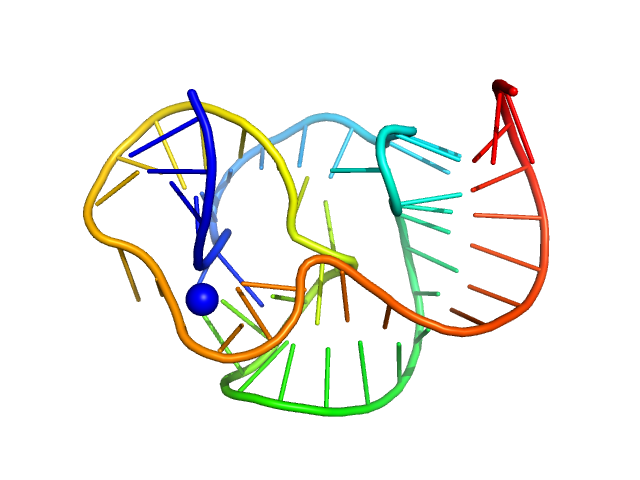
|
| Sample: |
ST9 WT (59 nt RNA) monomer, 19 kDa RNA
|
| Buffer: |
50 mM Tris pH 7.5, 100 mM NaCl, 1 mM DTT, 10 mM MgCl2, 0.0002% w/v NaN3, pH: |
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2024 Jan 30
|
A conserved viral RNA fold enables nuclease resistance across kingdoms of life.
Nucleic Acids Res 53(16) (2025)
Gezelle JG, Korn SM, McDonald JT, Gong Z, Erickson A, Huang CH, Yang F, Cronin M, Kuo YW, Wimberly BT, Steckelberg AL
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.7 |
nm |
| VolumePorod |
24 |
nm3 |
|
|
|
|
|
|
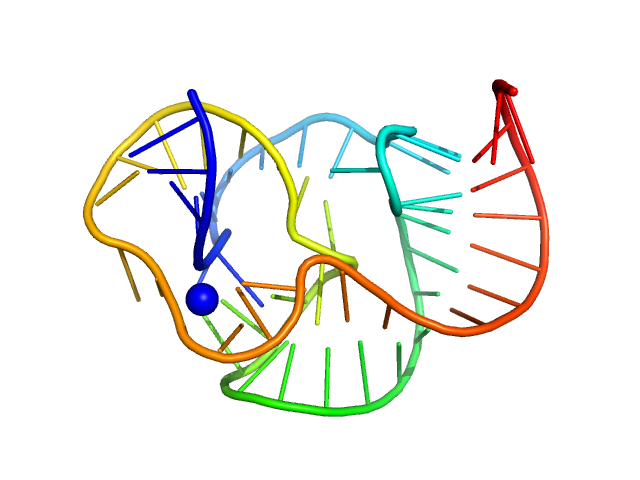
|
| Sample: |
ST9 WT (59 nt RNA) monomer, 19 kDa RNA
|
| Buffer: |
50 mM Tris pH 7.5, 100 mM NaCl, 1 mM DTT, 0.0002% w/v NaN3, pH: |
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2024 Jan 30
|
A conserved viral RNA fold enables nuclease resistance across kingdoms of life.
Nucleic Acids Res 53(16) (2025)
Gezelle JG, Korn SM, McDonald JT, Gong Z, Erickson A, Huang CH, Yang F, Cronin M, Kuo YW, Wimberly BT, Steckelberg AL
|
| RgGuinier |
2.1 |
nm |
| Dmax |
8.3 |
nm |
| VolumePorod |
26 |
nm3 |
|
|