|
|
|
|
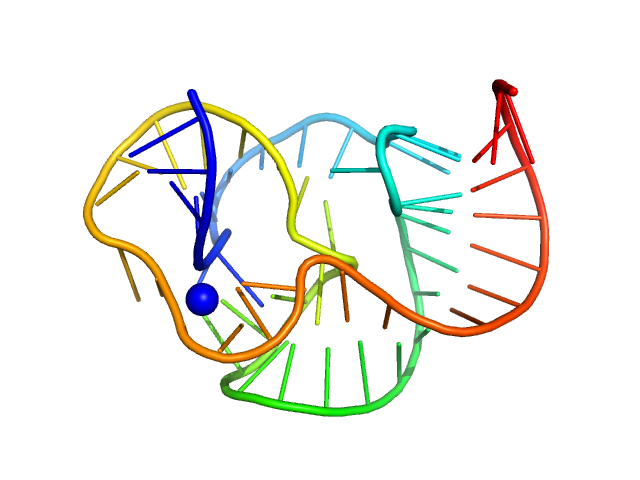
|
| Sample: |
ST9 PKmutant (59 nt RNA) monomer, 19 kDa RNA
|
| Buffer: |
50 mM Tris pH 7.5, 100 mM NaCl, 1 mM DTT, 0.0002% w/v NaN3, pH: |
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2024 Jan 30
|
A conserved viral RNA fold enables nuclease resistance across kingdoms of life.
Nucleic Acids Res 53(16) (2025)
Gezelle JG, Korn SM, McDonald JT, Gong Z, Erickson A, Huang CH, Yang F, Cronin M, Kuo YW, Wimberly BT, Steckelberg AL
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.8 |
nm |
| VolumePorod |
29 |
nm3 |
|
|
|
|
|
|
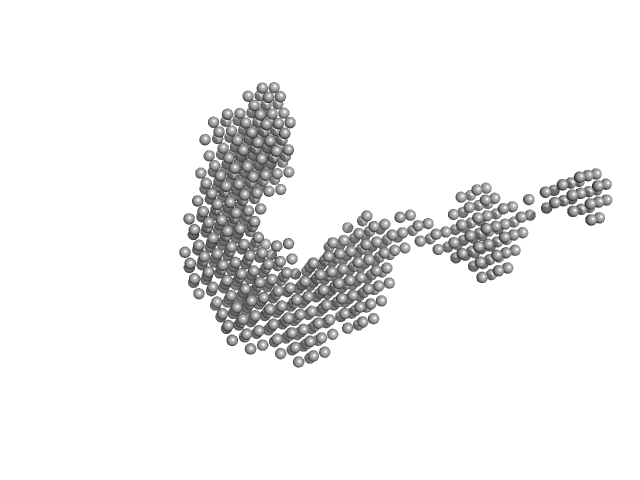
|
| Sample: |
E3 ubiquitin-protein ligase PRT1 monomer, 46 kDa Arabidopsis thaliana protein
|
| Buffer: |
50 mM Tris-HCl, 150 mM NaCl, 1 mM TCEP, 2% w/v glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2018 May 22
|
Structural basis for the recognition and ubiquitylation of type-2 N-degron substrate by PRT1 plant N-recognin
Nature Communications 16(1) (2025)
Yang W, Kim S, Kim M, Shin H, Lee J, Sandmann A, Park O, Dissmeyer N, Song H
|
| RgGuinier |
3.4 |
nm |
| Dmax |
12.7 |
nm |
| VolumePorod |
61 |
nm3 |
|
|
|
|
|
|
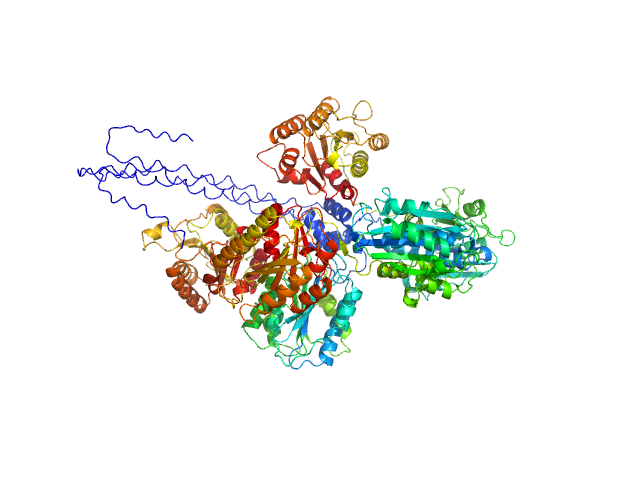
|
| Sample: |
Collagen alpha-1(VI) chain, 52 kDa Homo sapiens protein
Collagen alpha-2(VI) chain (R680H, K966N, Q967E), 53 kDa Homo sapiens protein
Collagen alpha-3(VI) chain (D2357R, K2367R, D2431V, R2441T, R2609A, R2610A), 54 kDa Homo sapiens protein
|
| Buffer: |
Tris-buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 Jul 27
|
Collagen VI microfibril structure reveals mechanism for molecular assembly and clustering of inherited pathogenic mutations.
Nat Commun 16(1):7549 (2025)
Godwin ARF, Becker MH, Dajani R, Snee M, Roseman AM, Baldock C
|
| RgGuinier |
4.8 |
nm |
| Dmax |
18.7 |
nm |
| VolumePorod |
478 |
nm3 |
|
|
|
|
|
|
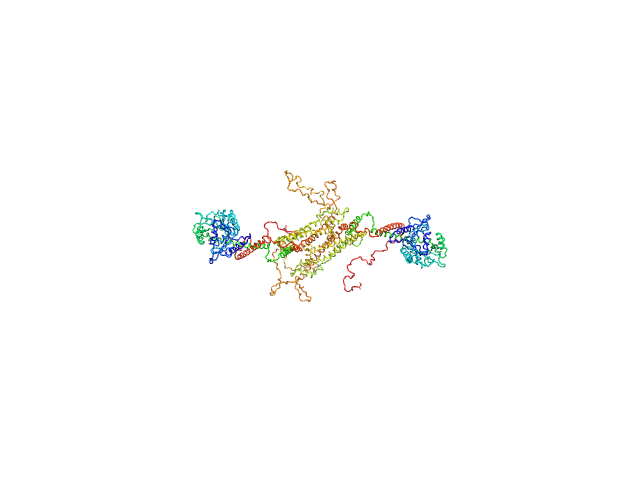
|
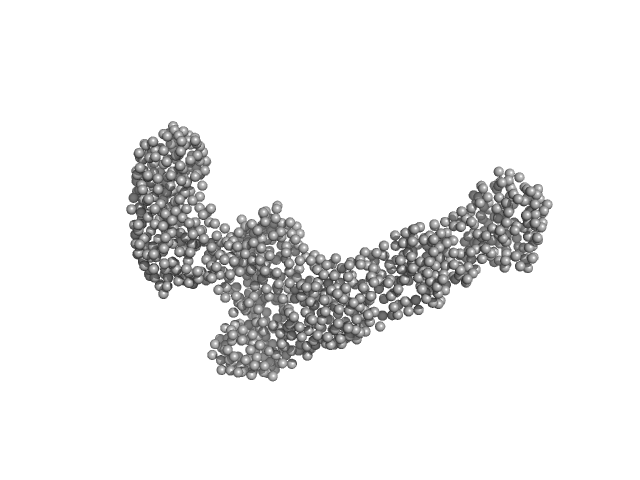
| Sample: |
Dynamin central region family protein dimer, 165 kDa Leishmania donovani protein
|
| Buffer: |
20 mM HEPES, 200 mM NaCl, 3% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2024 Apr 4
|
Biophysical analysis of an oligomerization-attenuated variant of the Leishmania donovani dynamin-1-like protein.
Mol Biochem Parasitol 263:111691 (2025)
Wuyts E, Sundaramoorthy R, Tulloch L, Monsieurs P, Eadsforth TC, Pintelon I, Timmermans JP, Dujardin JC, Domagalska MA, Caljon G, De Rycker M, Postis VLG, Wyllie S, Sterckx YG
|
| RgGuinier |
6.6 |
nm |
| Dmax |
25.0 |
nm |
| VolumePorod |
340 |
nm3 |
|
|
|
|
|
|
|
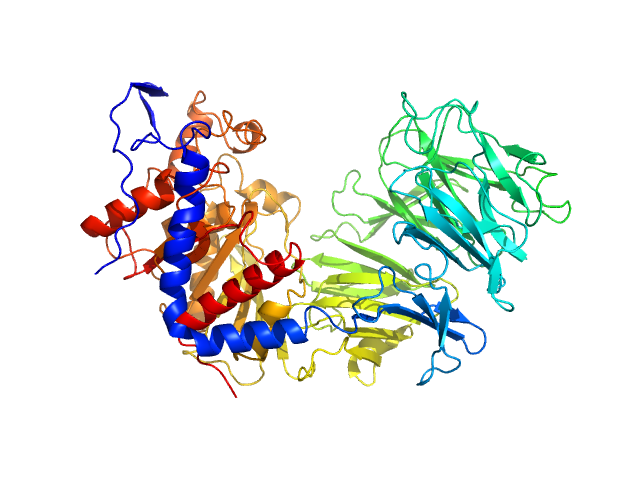
| Sample: |
Chaperone protein DnaK, 69 kDa Escherichia coli (strain … protein
Heme ligase, 28 kDa Plasmodium falciparum (isolate … protein
|
| Buffer: |
20 mM Tris-Cl, 200 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, Chemistry Division, Bhabha Atomic Research Centre on 2019 Feb 26
|
Recombinant expression and purification of Plasmodium heme detoxification protein in E. coli: Challenges and discoveries.
Protein Expr Purif 236:106789 (2025)
Singh R, Makde RD
|
| RgGuinier |
3.8 |
nm |
| Dmax |
13.8 |
nm |
| VolumePorod |
83 |
nm3 |
|
|
|
|
|
|
|
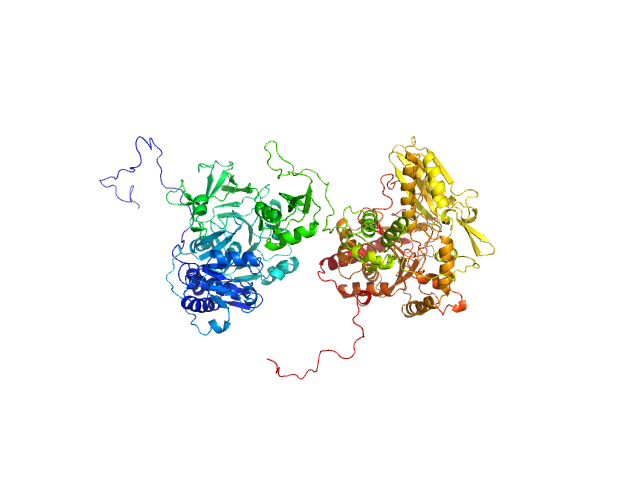
| Sample: |
Prolyl endopeptidase monomer, 78 kDa Trypanosoma cruzi protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Apr 24
|
Cryo-EM led analysis of open and closed conformations of Chagas vaccine candidate TcPOP.
Nat Commun 16(1):7164 (2025)
Batra S, Olmo F, Ragan TJ, Kaplan M, Calvaresi V, Frank AM, Lancey C, Assadipapari M, Ying C, Struwe WB, Hesketh EL, Kelly JM, Barfod L, Campeotto I
|
| RgGuinier |
3.0 |
nm |
| Dmax |
0.0 |
nm |
| VolumePorod |
142 |
nm3 |
|
|
|
|
|
|
|
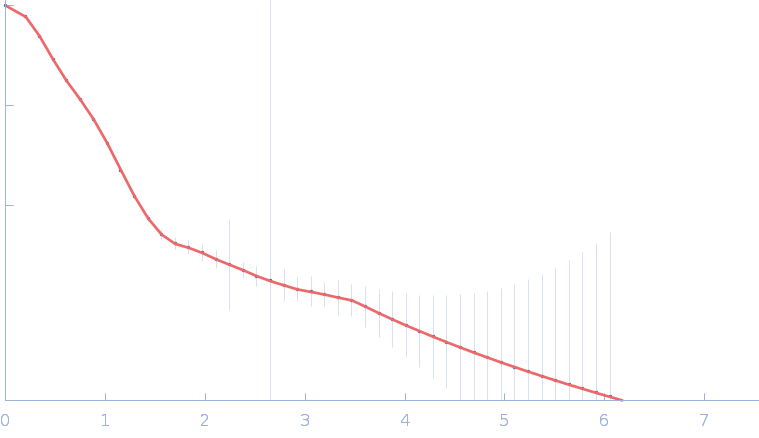
| Sample: |
Gramicidin S synthase 1 (W239S) monomer, 127 kDa Aneurinibacillus migulanus protein
|
| Buffer: |
20 mM Tris pH 7.5, 100 mM NaCl, 10 mM MgCl2, 2% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2023 Mar 14
|
Resting state of GrsA is a combination of two constitutively inactive isoforms that manifest long-range allosteric effects in presence of aminoacid and ATP
Raktim Roy
|
| RgGuinier |
4.1 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
187 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Gramicidin S synthase 1 (W239S) monomer, 126 kDa Brevibacillus brevis protein
|
| Buffer: |
20 mM Tris pH 7.5, 100 mM NaCl, 10 mM MgCl2, 2% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2023 Mar 14
|
Resting state of GrsA is a combination of two constitutively inactive isoforms that manifest long-range allosteric effects in presence of aminoacid and ATP
Raktim Roy
|
| RgGuinier |
4.3 |
nm |
| Dmax |
15.4 |
nm |
| VolumePorod |
186 |
nm3 |
|
|
|
|
|
|
|
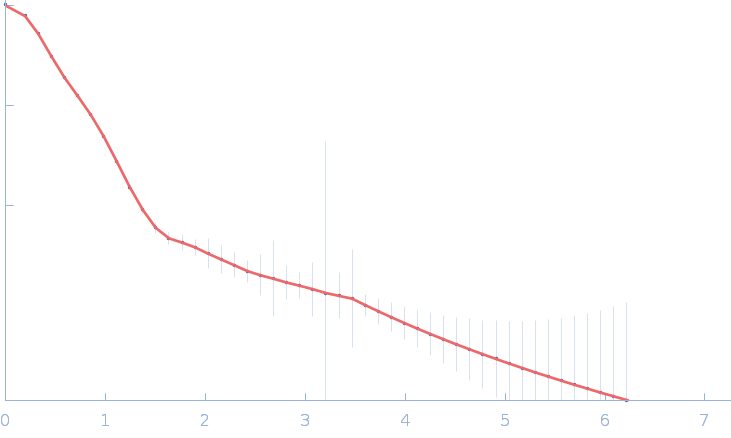
| Sample: |
Gramicidin S synthase 1 (W239S) monomer, 126 kDa Brevibacillus brevis protein
|
| Buffer: |
20 mM Tris pH 7.5, 100 mM NaCl, 10 mM MgCl2, 2% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2023 Mar 14
|
Resting state of GrsA is a combination of two constitutively inactive isoforms that manifest long-range allosteric effects in presence of aminoacid and ATP
Raktim Roy
|
| RgGuinier |
4.2 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
186 |
nm3 |
|
|
|
|
|
|
|
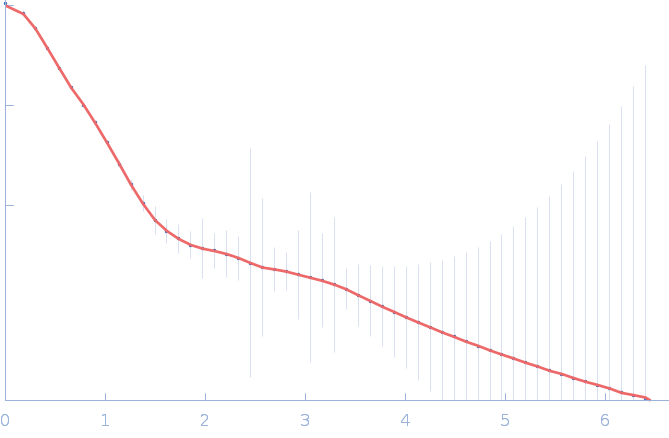
| Sample: |
Gramicidin S synthase 1 (W239S) monomer, 126 kDa Brevibacillus brevis protein
|
| Buffer: |
20 mM Tris pH 7.5, 100 mM NaCl, 10 mM MgCl2, 2% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2023 Mar 14
|
Resting state of GrsA is a combination of two constitutively inactive isoforms that manifest long-range allosteric effects in presence of aminoacid and ATP
Raktim Roy
|
| RgGuinier |
4.1 |
nm |
| Dmax |
17.5 |
nm |
| VolumePorod |
181 |
nm3 |
|
|