|
|
|
|
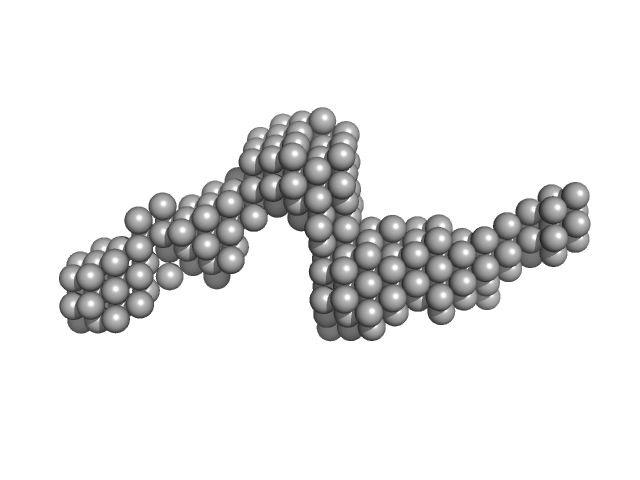
|
| Sample: |
Gli-55 - ssDNA aptamer specific to glioma brain tumor cells monomer, 19 kDa Artificially synthesized DNA
|
| Buffer: |
Dulbecco phosphate-buffered saline (DPBS), pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Oct 30
|
Development of DNA Aptamers for Visualization of Glial Brain Tumors and Detection of Circulating Tumor Cells
Molecular Therapy - Nucleic Acids (2023)
Kichkailo A, Narodov A, Komarova M, Zamay T, Zamay G, Kolovskaya O, Erakhtin E, Glazyrin Y, Veprintsev D, Moryachkov R, Zabluda V, Shchugoreva I, Artyushenko P, Mironov V, Morozov D, Khorzhevskii V, Gorbushin A, Koshmanova A, Nikolaeva E, Grinev I, Voronkovskii I, Grek D, Belugin K, Volzhentsev A, Badmaev O, Luzan N, Lukyanenko K, Peters G, Lapin I, Kirichenko A, Konarev P, Morozov E, Mironov G, Gargaun A, Muharemagic D, Zamay S, Kochkina E, Dymova M, Smolyarova T, Sokolov A, Modestov A, Tokarev N, Shepelevich N, Ozerskaya A, Chanchikova N, Krat A, Zukov R, Bakhtina V, Shnyakin P, Shesternya P, Svetlichnyi V, Petrova M, Artyukhov I, Tomilin F, Berezovski M
|
| RgGuinier |
2.8 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
34 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Gli-55 - ssDNA aptamer to glioma brain tumor cells monomer, 19 kDa synthetic construct DNA
|
| Buffer: |
Dulbecco phosphate-buffered saline (DPBS), pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Oct 30
|
Development of DNA Aptamers for Visualization of Glial Brain Tumors and Detection of Circulating Tumor Cells
Molecular Therapy - Nucleic Acids (2023)
Kichkailo A, Narodov A, Komarova M, Zamay T, Zamay G, Kolovskaya O, Erakhtin E, Glazyrin Y, Veprintsev D, Moryachkov R, Zabluda V, Shchugoreva I, Artyushenko P, Mironov V, Morozov D, Khorzhevskii V, Gorbushin A, Koshmanova A, Nikolaeva E, Grinev I, Voronkovskii I, Grek D, Belugin K, Volzhentsev A, Badmaev O, Luzan N, Lukyanenko K, Peters G, Lapin I, Kirichenko A, Konarev P, Morozov E, Mironov G, Gargaun A, Muharemagic D, Zamay S, Kochkina E, Dymova M, Smolyarova T, Sokolov A, Modestov A, Tokarev N, Shepelevich N, Ozerskaya A, Chanchikova N, Krat A, Zukov R, Bakhtina V, Shnyakin P, Shesternya P, Svetlichnyi V, Petrova M, Artyukhov I, Tomilin F, Berezovski M
|
| RgGuinier |
2.8 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
35 |
nm3 |
|
|
|
|
|
|
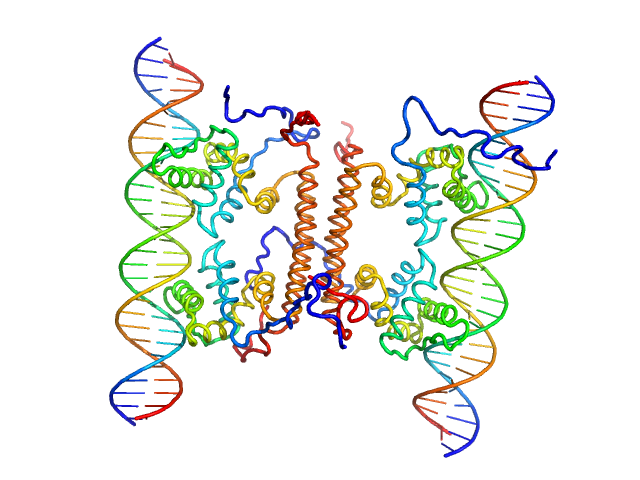
|
| Sample: |
YdaT_toxin domain-containing protein tetramer, 74 kDa Escherichia coli O157:H7 protein
Om 30 base pair dsDNA dimer, 37 kDa DNA
|
| Buffer: |
20 mM Tris-HCl, 200 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Apr 14
|
Structural basis of DNA binding by YdaT, a functional equivalent of the CII repressor in the cryptic prophage CP-933P from Escherichia coli
O157:H7
Acta Crystallographica Section D Structural Biology 79(3):245-258 (2023)
Prolič-Kalinšek M, Volkov A, Hadži S, Van Dyck J, Bervoets I, Charlier D, Loris R
|
| RgGuinier |
4.2 |
nm |
| Dmax |
12.8 |
nm |
| VolumePorod |
190 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Neural cell adhesion molecule L1 monomer, 24 kDa Homo sapiens protein
|
| Buffer: |
50 mM MES pH 6.5, 150 mM NaCl, 5% glycerol, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 May 11
|
X‐ray structure and function of fibronectin domains two and three of the neural cell adhesion molecule L1
The FASEB Journal 37(3) (2023)
Guédez G, Loers G, Jeffries C, Kozak S, Meijers R, Svergun D, Schachner M, Löw C
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
68 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Neural cell adhesion molecule L1 monomer, 24 kDa Homo sapiens protein
|
| Buffer: |
50 mM MES pH 6.5, 150 mM NaCl, 5% glycerol, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 May 11
|
X‐ray structure and function of fibronectin domains two and three of the neural cell adhesion molecule L1
The FASEB Journal 37(3) (2023)
Guédez G, Loers G, Jeffries C, Kozak S, Meijers R, Svergun D, Schachner M, Löw C
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.9 |
nm |
| VolumePorod |
72 |
nm3 |
|
|
|
|
|
|
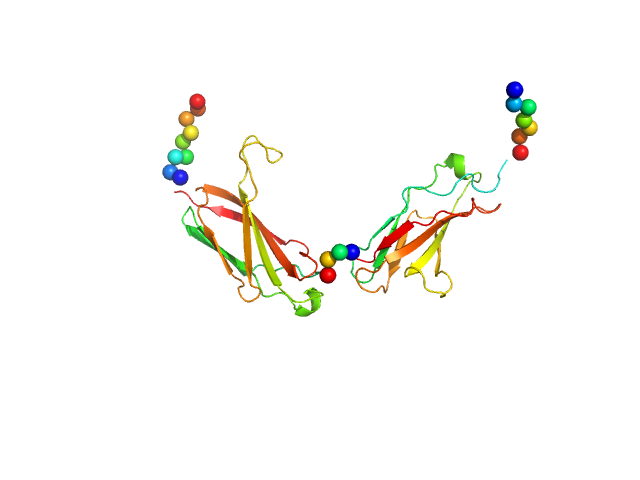
|
| Sample: |
Neural cell adhesion molecule L1 monomer, 24 kDa Homo sapiens protein
|
| Buffer: |
50 mM MES pH 6.5, 150 mM NaCl, 5% glycerol, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 May 11
|
X‐ray structure and function of fibronectin domains two and three of the neural cell adhesion molecule L1
The FASEB Journal 37(3) (2023)
Guédez G, Loers G, Jeffries C, Kozak S, Meijers R, Svergun D, Schachner M, Löw C
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.6 |
nm |
| VolumePorod |
72 |
nm3 |
|
|
|
|
|
|
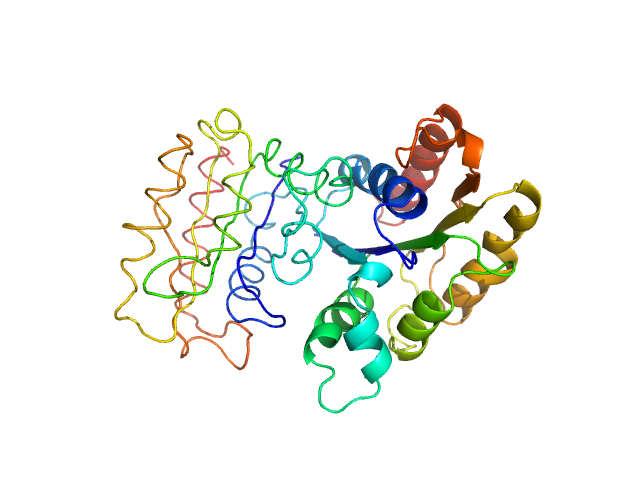
|
| Sample: |
NADPH-dependent FMN reductase dimer, 41 kDa Paracoccus denitrificans protein
|
| Buffer: |
50 mM sodium phosphate buffer, 300 mM NaCl, 500 mM imidazole, pH: 8 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, CEITEC on 2016 Feb 22
|
Structural Insight into Catalysis by the Flavin-Dependent NADH Oxidase (Pden_5119) of Paracoccus denitrificans.
Int J Mol Sci 24(4) (2023)
Kryl M, Sedláček V, Kučera I
|
| RgGuinier |
2.3 |
nm |
| Dmax |
6.3 |
nm |
| VolumePorod |
70 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
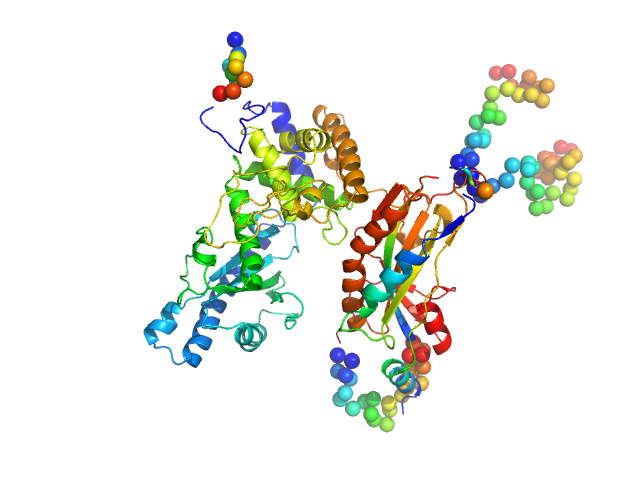
NURS complex subunit red1 monomer, 8 kDa Schizosaccharomyces pombe (strain … protein
Poly(A) polymerase pla1 monomer, 65 kDa Schizosaccharomyces pombe (strain … protein
|
| Buffer: |
20 mM HEPES, pH 7.5, 150 mM NaCl, 1mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Nov 12
|
Mechanistic insights into RNA surveillance by the canonical poly(A) polymerase Pla1 of the MTREC complex.
Nat Commun 14(1):772 (2023)
Soni K, Sivadas A, Horvath A, Dobrev N, Hayashi R, Kiss L, Simon B, Wild K, Sinning I, Fischer T
|
| RgGuinier |
3.3 |
nm |
| Dmax |
10.7 |
nm |
| VolumePorod |
138 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
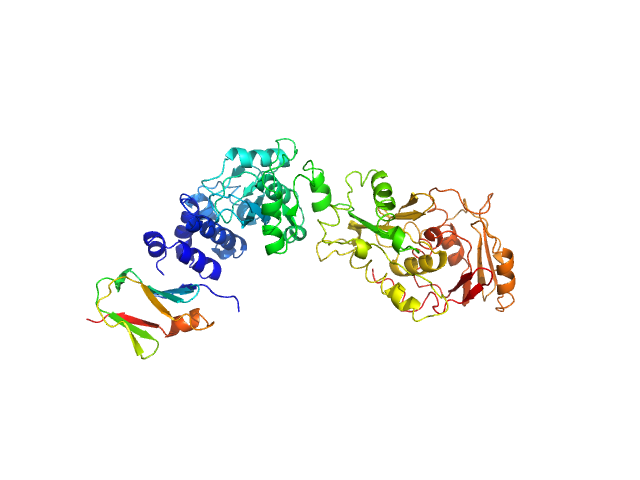
SAVED domain-containing protein monomer, 58 kDa Sulfurihydrogenibium sp. (strain … protein
Uncharacterized protein (putative MazF-like toxin) monomer, 9 kDa Sulfurihydrogenibium sp. (strain … protein
|
| Buffer: |
20 mM Tris, 50 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Dec 14
|
Antiviral signalling by a cyclic nucleotide activated CRISPR protease.
Nature 614(7946):168-174 (2023)
Rouillon C, Schneberger N, Chi H, Blumenstock K, Da Vela S, Ackermann K, Moecking J, Peter MF, Boenigk W, Seifert R, Bode BE, Schmid-Burgk JL, Svergun D, Geyer M, White MF, Hagelueken G
|
| RgGuinier |
3.4 |
nm |
| Dmax |
11.7 |
nm |
| VolumePorod |
93 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Ac-(POG)4-QG-(POG)5-NH2 trimer, 11 kDa synthetic construct protein
|
| Buffer: |
20 mM L-histidine, 138 mM NaCl, 2.7 mM KCL,, pH: 6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Feb 1
|
A solution structure analysis reveals a bent collagen triple helix in the complement activation recognition molecule mannan-binding lectin.
J Biol Chem 299(2):102799 (2023)
Iqbal H, Fung KW, Gor J, Bishop AC, Makhatadze GI, Brodsky B, Perkins SJ
|
| RgGuinier |
2.1 |
nm |
| Dmax |
8.7 |
nm |
| VolumePorod |
7 |
nm3 |
|
|