|
|
|
|
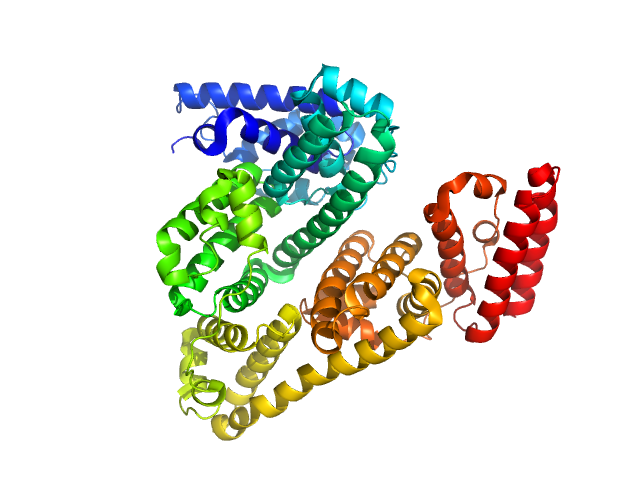
|
| Sample: |
Bovine serum albumin monomer, 66 kDa Bos taurus protein
|
| Buffer: |
TRIS 50mM, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Dec 21
|
A high flux setup for millisecond-scale small-angle X-ray scattering studies on macromolecular solutions
Clement Blanchet
|
|
|
|
|
|
|
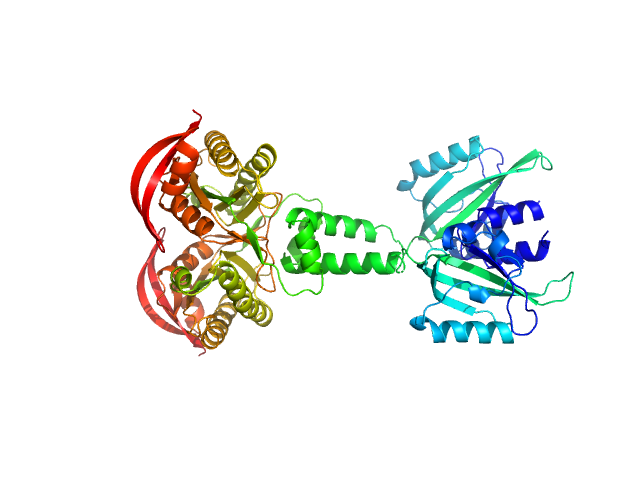
|
| Sample: |
PAS fold family dimer, 78 kDa Coleofasciculus chthonoplastes PCC … protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 5 mM MgCl2, 5 % w/v Glycerol, 1 mM ApCpp, pH: 7.5 |
| Experiment: |
SAXS
data collected at cSAXS, Swiss Light Source on 2015 Mar 11
|
MPAC Delta132
Robert Lindner
|
| RgGuinier |
4.0 |
nm |
| Dmax |
12.8 |
nm |
| VolumePorod |
99 |
nm3 |
|
|
|
|
|
|
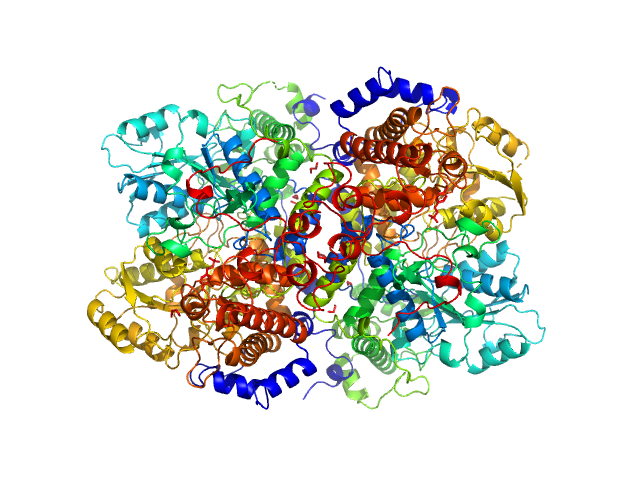
|
| Sample: |
Glycine decarboxylase dimer, 219 kDa Homo sapiens protein
|
| Buffer: |
10 mM TRIS pH 7.5, 100 mM NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Nov 12
|
Glycine decarboxylase (P-protein of glycine cleavage system)
Bart Van Laer
|
| RgGuinier |
3.9 |
nm |
| VolumePorod |
304 |
nm3 |
|
|
|
|
|
|
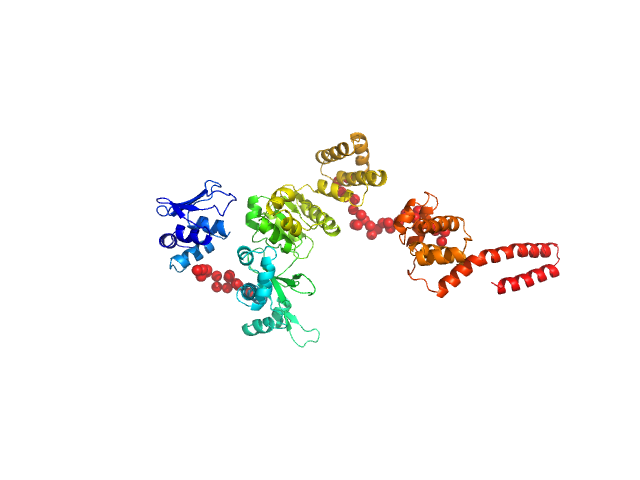
|
| Sample: |
DNA primase monomer, 69 kDa Bacillus subtilis protein
|
| Buffer: |
25 mM Tris-HCl,100 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2018 Oct 1
|
Full length of DnaG primase from Bacillus subtilis
Zhongchuan Liu
|
| RgGuinier |
3.7 |
nm |
| Dmax |
14.5 |
nm |
| VolumePorod |
119 |
nm3 |
|
|
|
|
|
|
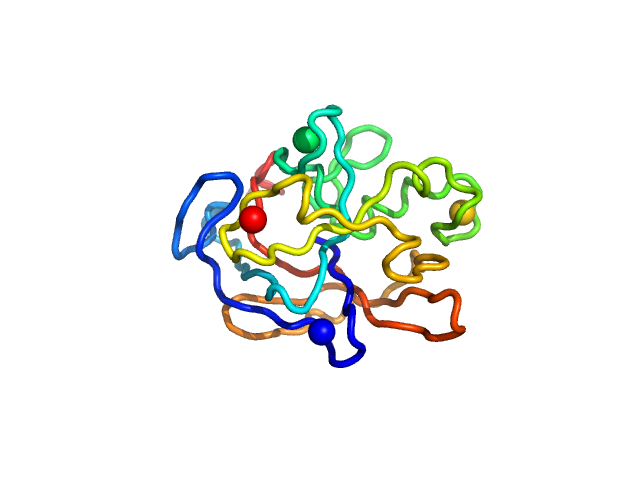
|
| Sample: |
Iron-regulated protein FrpC (amino acids 415-591) monomer, 19 kDa Neisseria meningitidis protein
|
| Buffer: |
5 mM Tris 50 mM NaCl 10 mM CaCl2 0.1% NaN3, pH: 7.4 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, CEITEC on 2013 Oct 3
|
Self-processing module of Iron-regulated protein FrpC
Vojtěch Kubáň
|
| RgGuinier |
1.7 |
nm |
| Dmax |
4.5 |
nm |
| VolumePorod |
21 |
nm3 |
|
|
|
|
|
|
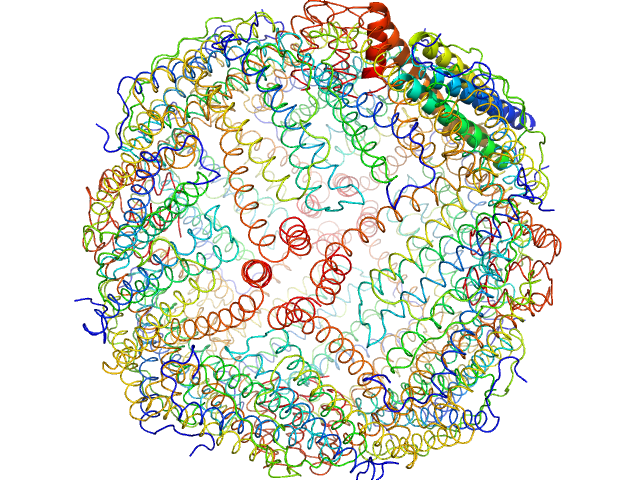
|
| Sample: |
Horse spleen apoferritin 24-mer, 476 kDa Equus caballus protein
|
| Buffer: |
tbs, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jul 15
|
WAXS benchmark on standard proteins
Maxim Petoukhov
|
|
|
|
|
|
|
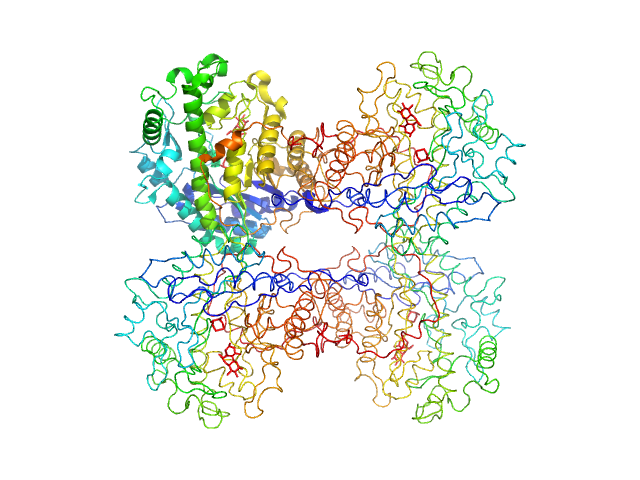
|
| Sample: |
Beta-amylase tetramer, 224 kDa Ipomoea batatas protein
|
| Buffer: |
tbs, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jul 15
|
WAXS benchmark on standard proteins
Maxim Petoukhov
|
|
|
|
|
|
|
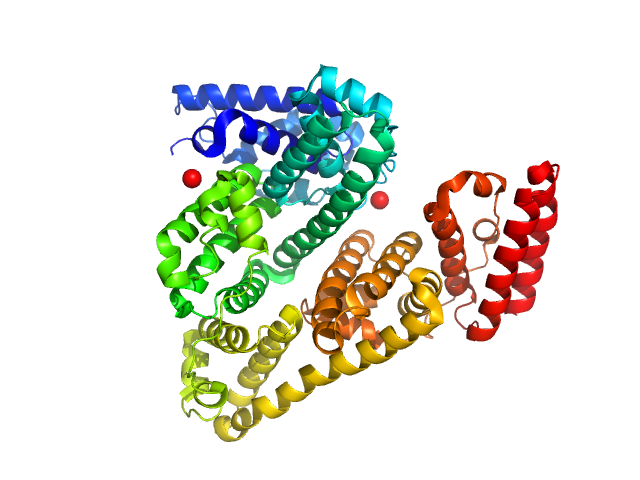
|
| Sample: |
Serum albumin monomer, 66 kDa Bos taurus protein
|
| Buffer: |
Hepes, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jun 25
|
WAXS benchmark on standard proteins
Maxim Petoukhov
|
|
|
|
|
|
|
|
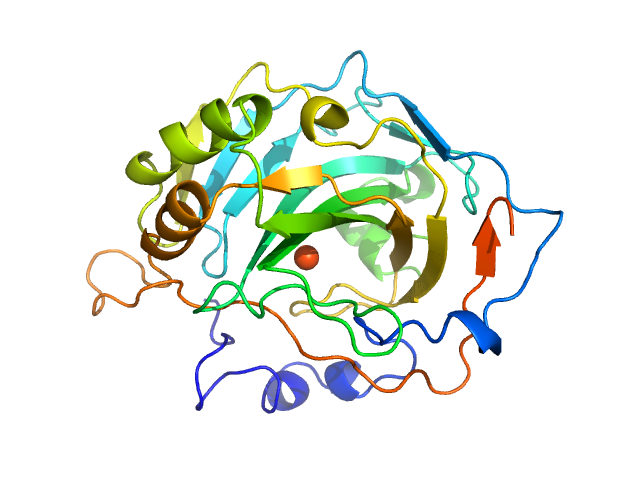
| Sample: |
Carbonic anhydrase 2 monomer, 29 kDa Bos taurus protein
|
| Buffer: |
Tbs, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jul 16
|
WAXS benchmark on standard proteins
Maxim Petoukhov
|
|
|
|
|
|
|
|
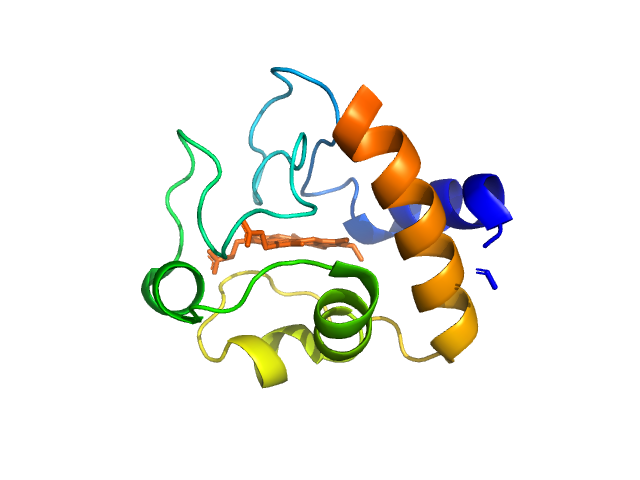
| Sample: |
Cytochrome c monomer, 12 kDa Equus caballus protein
|
| Buffer: |
tbs, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jul 16
|
WAXS benchmark on standard proteins
Maxim Petoukhov
|
|
|