|
|
|
|
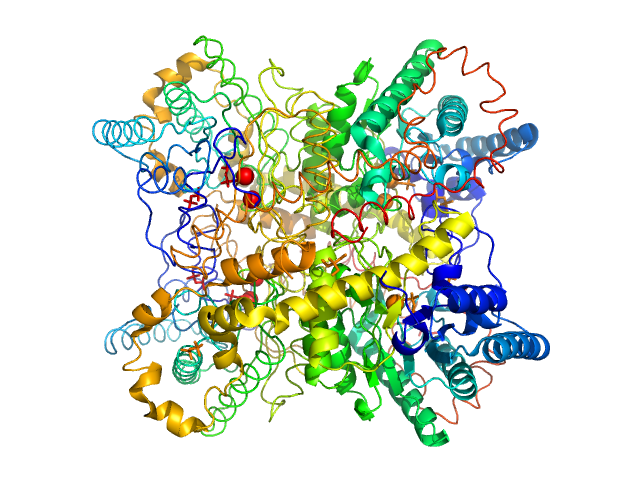
|
| Sample: |
Xylose isomerase tetramer, 172 kDa Streptomyces rubiginosus protein
|
| Buffer: |
100 mM tris pH 8.0, 100 mM NaCl, 1 mM MgCl2, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Oct 17
|
WAXS benchmark on standard proteins
Maxim Petoukhov
|
|
|
|
|
|
|
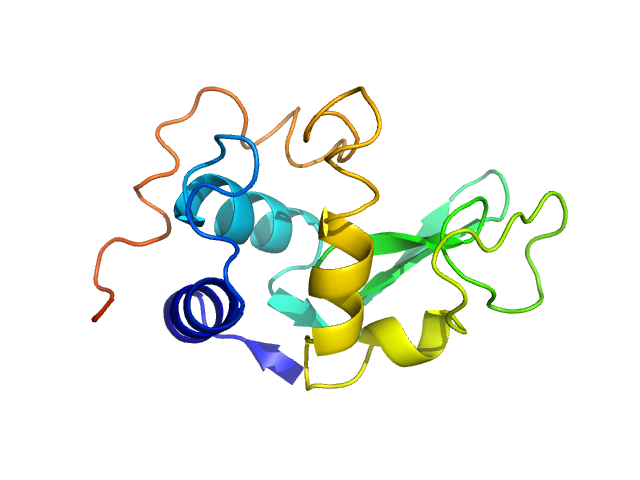
|
| Sample: |
Lysozyme C monomer, 14 kDa Gallus gallus protein
|
| Buffer: |
40 mM Sodium Acetate pH 4.0, 50 mM NaCl, pH: 4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 May 31
|
WAXS benchmark on standard proteins
Maxim Petoukhov
|
|
|
|
|
|
|
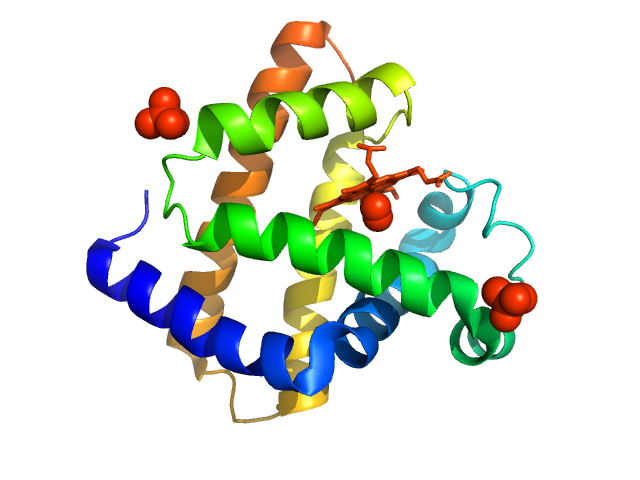
|
| Sample: |
Myoglobin monomer, 17 kDa Equus caballus protein
|
| Buffer: |
100 mM tris pH 7.5, 100 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jun 2
|
WAXS benchmark on standard proteins
Maxim Petoukhov
|
|
|
|
|
|
|
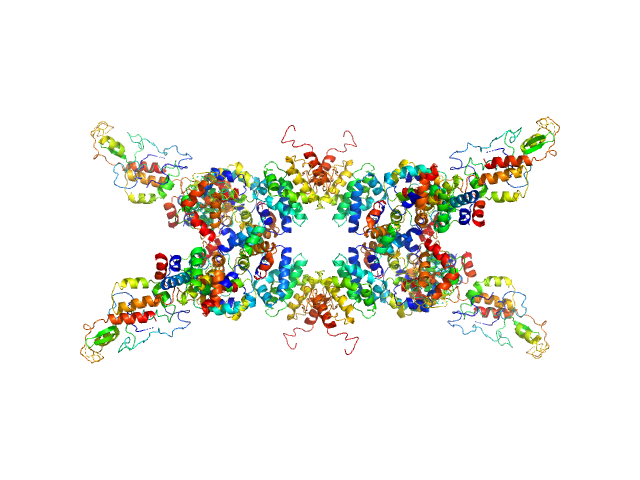
|
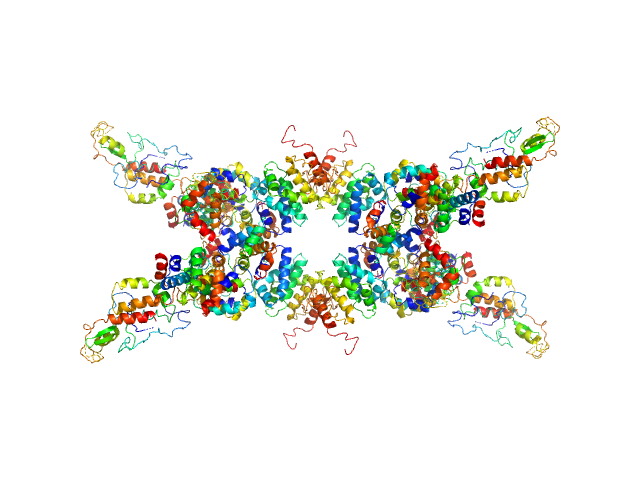
| Sample: |
Senescence-associated E3 ubiquitin ligase 1 tetramer, 355 kDa Arabidopsis thaliana protein
|
| Buffer: |
50 mM Tris, 250 mM NaCl, pH: 9 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jun 2
|
Senescence-associated ubiquitin ligase 1 (SAUL1)
Haifa El Kilani,
Al Kikhney
|
| RgGuinier |
6.5 |
nm |
| Dmax |
24.0 |
nm |
|
|
|
|
|
|
|
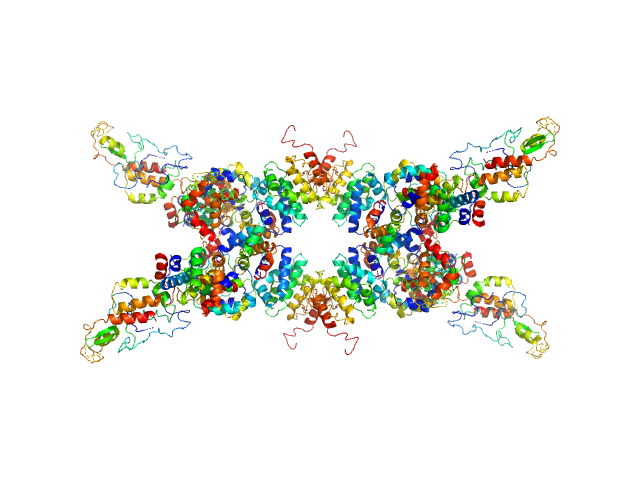
| Sample: |
Senescence-associated E3 ubiquitin ligase 1 tetramer, 355 kDa Arabidopsis thaliana protein
|
| Buffer: |
50 mM Tris, 250 mM NaCl, pH: 9 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jun 2
|
Senescence-associated ubiquitin ligase 1 (SAUL1)
Haifa El Kilani,
Al Kikhney
|
| RgGuinier |
5.9 |
nm |
| Dmax |
22.0 |
nm |
|
|
|
|
|
|
|
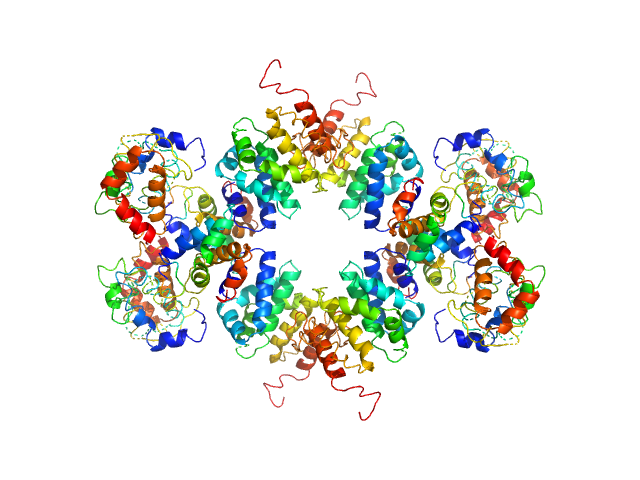
| Sample: |
Senescence-associated E3 ubiquitin ligase 1 tetramer, 355 kDa Arabidopsis thaliana protein
|
| Buffer: |
50 mM Tris, 250 mM NaCl, pH: 9 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jun 2
|
Senescence-associated ubiquitin ligase 1 (SAUL1)
Haifa El Kilani,
Al Kikhney
|
| RgGuinier |
5.9 |
nm |
| Dmax |
23.0 |
nm |
|
|
|
|
|
|
|
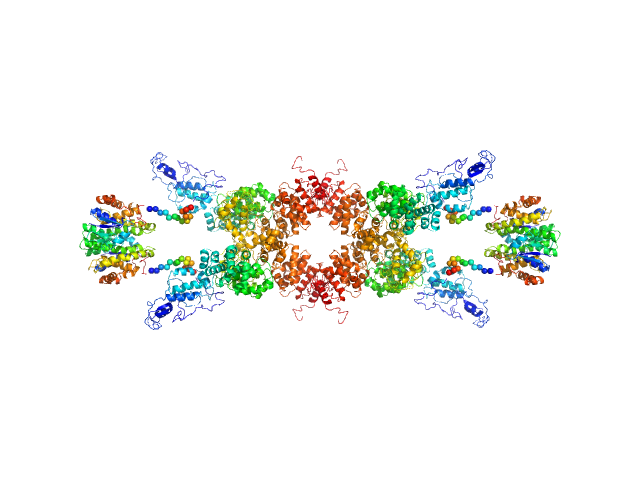
| Sample: |
U-box domain-containing protein 44 tetramer, 185 kDa Arabidopsis thaliana protein
|
| Buffer: |
50 mM Tris, 250 mM NaCl, pH: 9 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jun 2
|
Senescence-associated ubiquitin ligase 1 (SAUL1)
Haifa El Kilani,
Al Kikhney
|
| RgGuinier |
4.5 |
nm |
| Dmax |
17.4 |
nm |
| VolumePorod |
245 |
nm3 |
|
|
|
|
|
|
|
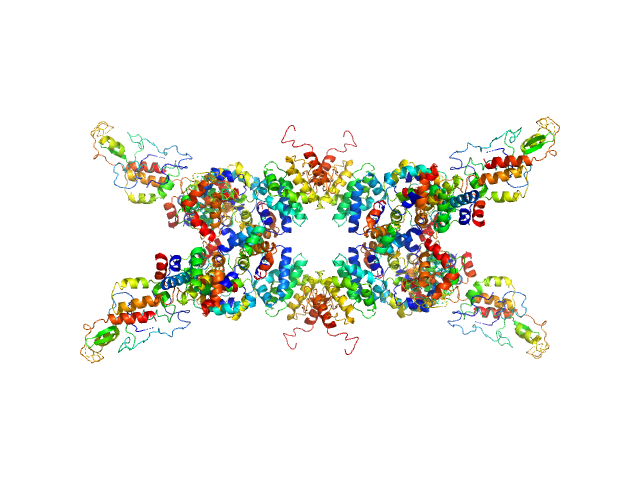
| Sample: |
Senescence-associated E3 ubiquitin ligase 1 tetramer, 355 kDa Arabidopsis thaliana protein
Glutathione S-transferase tetramer, 106 kDa Schistosoma japonicum protein
|
| Buffer: |
50 mM Tris, 250 mM NaCl, pH: 9 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2014 Jan 20
|
Senescence-associated ubiquitin ligase 1 (SAUL1)
Haifa El Kilani,
Al Kikhney
|
| RgGuinier |
7.6 |
nm |
| Dmax |
28.0 |
nm |
| VolumePorod |
778 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Senescence-associated E3 ubiquitin ligase 1 tetramer, 355 kDa Arabidopsis thaliana protein
|
| Buffer: |
50 mM Tris, 250 mM NaCl, pH: 9 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jun 2
|
Senescence-associated ubiquitin ligase 1 (SAUL1)
Haifa El Kilani,
Al Kikhney
|
| RgGuinier |
4.6 |
nm |
| Dmax |
17.2 |
nm |
| VolumePorod |
230 |
nm3 |
|
|
|
|
|
|
|
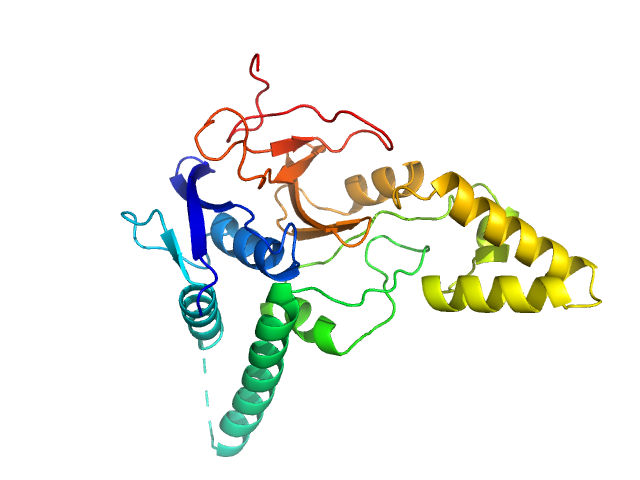
| Sample: |
Ubiquitin carboxyl-terminal hydrolase MINDY-2 monomer, 31 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 5 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Nov 23
|
Mechanism of activation and regulation of deubiquitinase activity in MINDY1 and MINDY2.
Mol Cell (2021)
Abdul Rehman SA, Armstrong LA, Lange SM, Kristariyanto YA, Gräwert TW, Knebel A, Svergun DI, Kulathu Y
|
| RgGuinier |
2.1 |
nm |
| Dmax |
6.5 |
nm |
| VolumePorod |
45 |
nm3 |
|
|