|
|
|
|
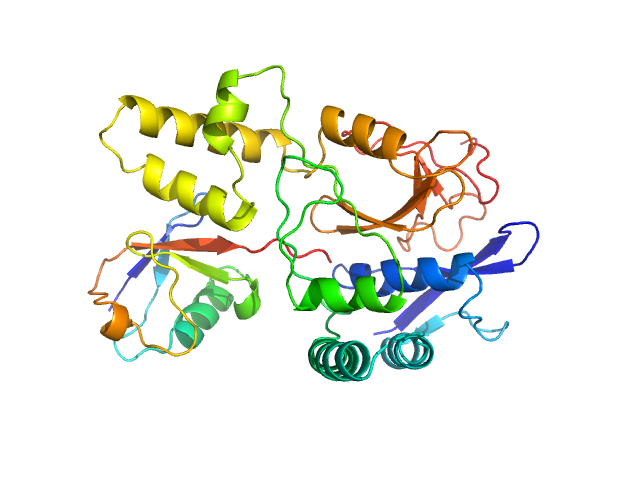
|
| Sample: |
Ubiquitin carboxyl-terminal hydrolase MINDY-2 monomer, 31 kDa Homo sapiens protein
Polyubiquitin-C monomer, 9 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 5 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Nov 23
|
Mechanism of activation and regulation of deubiquitinase activity in MINDY1 and MINDY2.
Mol Cell (2021)
Abdul Rehman SA, Armstrong LA, Lange SM, Kristariyanto YA, Gräwert TW, Knebel A, Svergun DI, Kulathu Y
|
| RgGuinier |
2.3 |
nm |
| Dmax |
6.8 |
nm |
| VolumePorod |
58 |
nm3 |
|
|
|
|
|
|
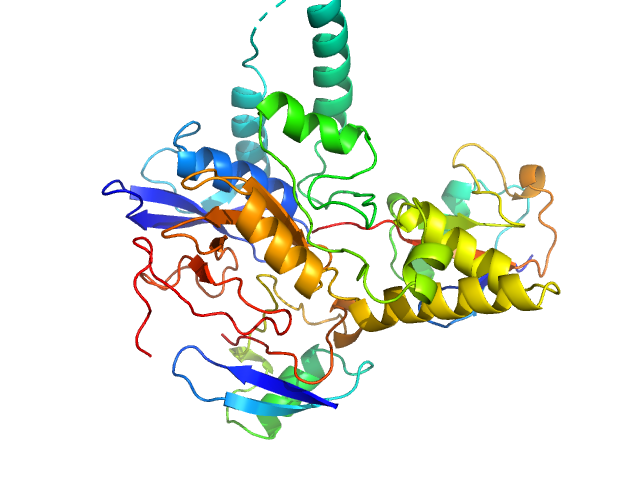
|
| Sample: |
Ubiquitin carboxyl-terminal hydrolase C266A mutant monomer, 31 kDa Homo sapiens protein
Polyubiquitin-C dimer, 17 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 5 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Nov 23
|
Mechanism of activation and regulation of deubiquitinase activity in MINDY1 and MINDY2.
Mol Cell (2021)
Abdul Rehman SA, Armstrong LA, Lange SM, Kristariyanto YA, Gräwert TW, Knebel A, Svergun DI, Kulathu Y
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
68 |
nm3 |
|
|
|
|
|
|
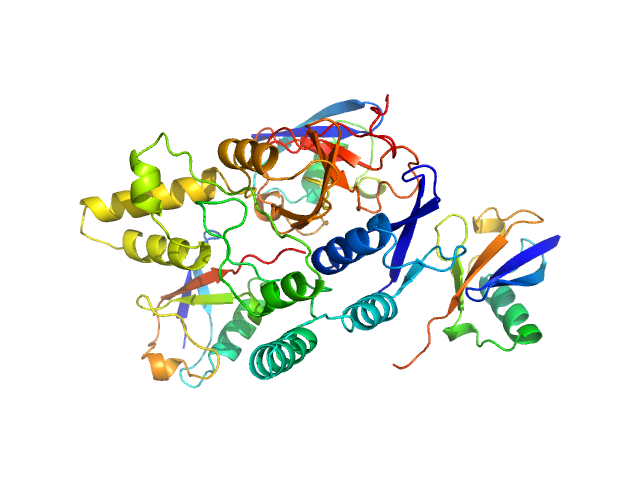
|
| Sample: |
Ubiquitin carboxyl-terminal hydrolase C266A mutant monomer, 31 kDa Homo sapiens protein
Polyubiquitin-C trimer, 26 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 5 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 May 26
|
Mechanism of activation and regulation of deubiquitinase activity in MINDY1 and MINDY2.
Mol Cell (2021)
Abdul Rehman SA, Armstrong LA, Lange SM, Kristariyanto YA, Gräwert TW, Knebel A, Svergun DI, Kulathu Y
|
| RgGuinier |
2.8 |
nm |
| Dmax |
8.8 |
nm |
| VolumePorod |
97 |
nm3 |
|
|
|
|
|
|
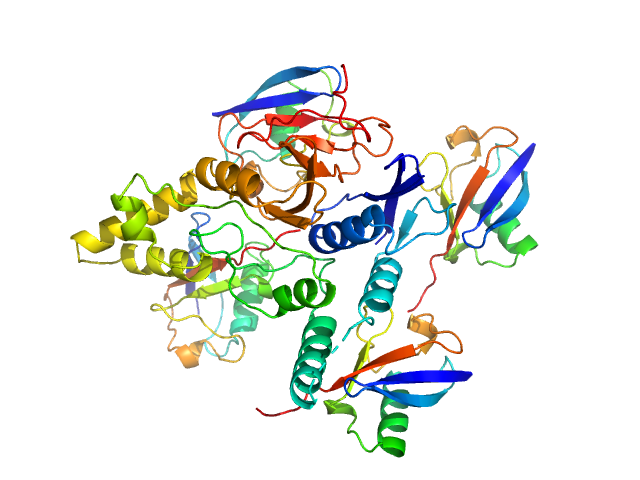
|
| Sample: |
Ubiquitin carboxyl-terminal hydrolase C266A mutant monomer, 31 kDa Homo sapiens protein
Polyubiquitin-C tetramer, 34 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 5 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 May 26
|
Mechanism of activation and regulation of deubiquitinase activity in MINDY1 and MINDY2.
Mol Cell (2021)
Abdul Rehman SA, Armstrong LA, Lange SM, Kristariyanto YA, Gräwert TW, Knebel A, Svergun DI, Kulathu Y
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.2 |
nm |
| VolumePorod |
111 |
nm3 |
|
|
|
|
|
|
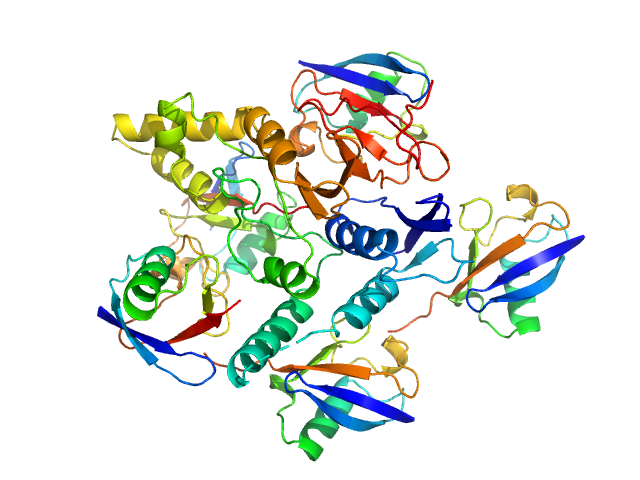
|
| Sample: |
Ubiquitin carboxyl-terminal hydrolase C266A mutant monomer, 31 kDa Homo sapiens protein
Polyubiquitin-C pentamer, 43 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 5 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 May 26
|
Mechanism of activation and regulation of deubiquitinase activity in MINDY1 and MINDY2.
Mol Cell (2021)
Abdul Rehman SA, Armstrong LA, Lange SM, Kristariyanto YA, Gräwert TW, Knebel A, Svergun DI, Kulathu Y
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
116 |
nm3 |
|
|
|
|
|
|
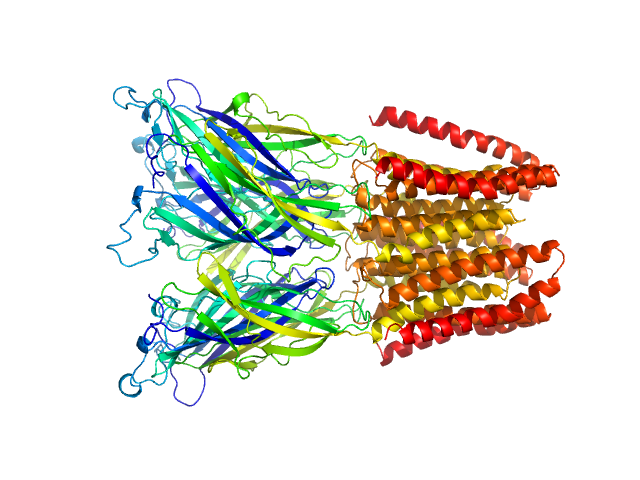
|
| Sample: |
Proton-gated ion channel pentamer, 183 kDa Gloeobacter violaceus (strain … protein
|
| Buffer: |
D2O, 20 mM Tris, 150 mM NaCl, 0.5 mM matched-out deuterated DDM,, pH: 7.5 |
| Experiment: |
SANS
data collected at D22, Institut Laue-Langevin (ILL) on 2020 Aug 22
|
Probing solution structure of the pentameric ligand-gated ion channel GLIC by small-angle neutron scattering
Proceedings of the National Academy of Sciences 118(37):e2108006118 (2021)
Lycksell M, Rovšnik U, Bergh C, Johansen N, Martel A, Porcar L, Arleth L, Howard R, Lindahl E
|
| RgGuinier |
3.8 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
274 |
nm3 |
|
|
|
|
|
|
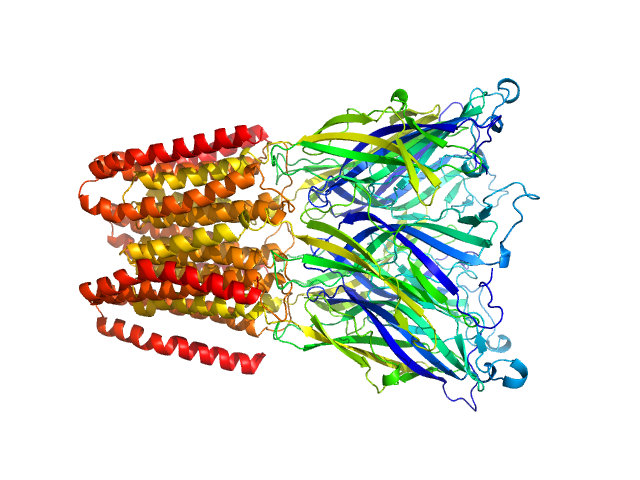
|
| Sample: |
Proton-gated ion channel pentamer, 183 kDa Gloeobacter violaceus (strain … protein
|
| Buffer: |
D2O, 20 mM citrate, 150 mM NaCl, 0.5 mM match-out deuterated DDM, pH: 3 |
| Experiment: |
SANS
data collected at D22, Institut Laue-Langevin (ILL) on 2020 Aug 22
|
Probing solution structure of the pentameric ligand-gated ion channel GLIC by small-angle neutron scattering
Proceedings of the National Academy of Sciences 118(37):e2108006118 (2021)
Lycksell M, Rovšnik U, Bergh C, Johansen N, Martel A, Porcar L, Arleth L, Howard R, Lindahl E
|
| RgGuinier |
3.8 |
nm |
| Dmax |
12.7 |
nm |
| VolumePorod |
279 |
nm3 |
|
|
|
|
|
|
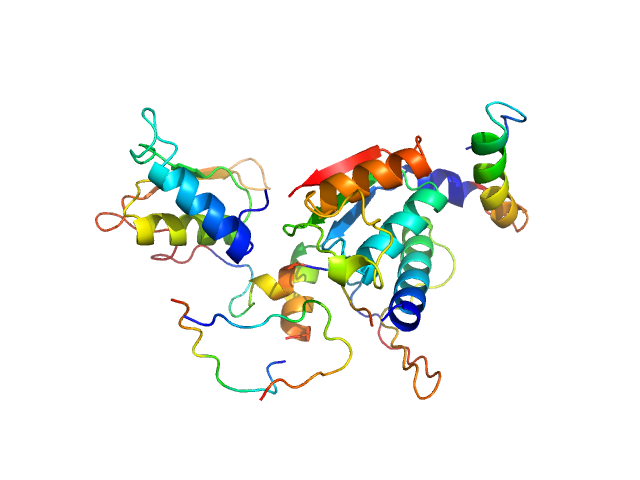
|
| Sample: |
Ubiquitin-like modifier-activating enzyme 5 dimer, 68 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Jun 13
|
Structure and dynamics of UBA5-UFM1 complex formation showing new insights in the UBA5 activation mechanism
Journal of Structural Biology :107796 (2021)
Fuchs S, Kikhney A, Schubert R, Kaiser C, Liebau E, Svergun D, Betzel C, Perbandt M
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
74 |
nm3 |
|
|
|
|
|
|
|
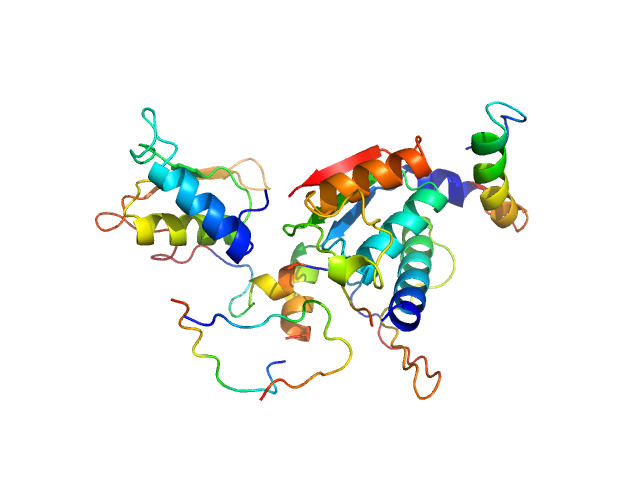
| Sample: |
Ubiquitin-like modifier-activating enzyme 5 dimer, 68 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Jun 13
|
Structure and dynamics of UBA5-UFM1 complex formation showing new insights in the UBA5 activation mechanism
Journal of Structural Biology :107796 (2021)
Fuchs S, Kikhney A, Schubert R, Kaiser C, Liebau E, Svergun D, Betzel C, Perbandt M
|
| RgGuinier |
2.9 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
82 |
nm3 |
|
|
|
|
|
|
|
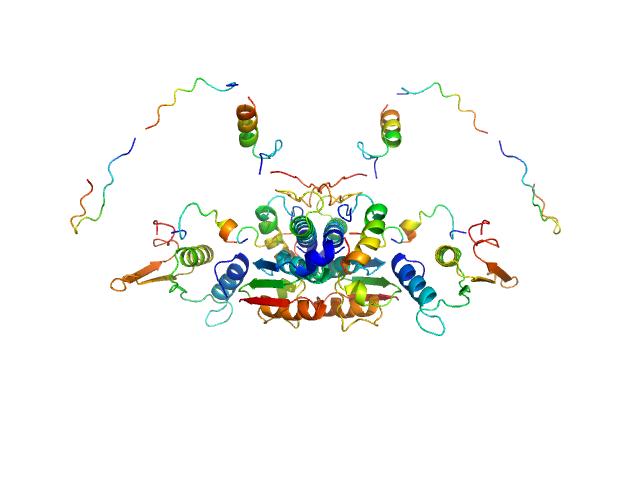
| Sample: |
Ubiquitin-like modifier-activating enzyme 5 dimer, 68 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Jun 13
|
Structure and dynamics of UBA5-UFM1 complex formation showing new insights in the UBA5 activation mechanism
Journal of Structural Biology :107796 (2021)
Fuchs S, Kikhney A, Schubert R, Kaiser C, Liebau E, Svergun D, Betzel C, Perbandt M
|
| RgGuinier |
3.2 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
108 |
nm3 |
|
|