|
|
|
|
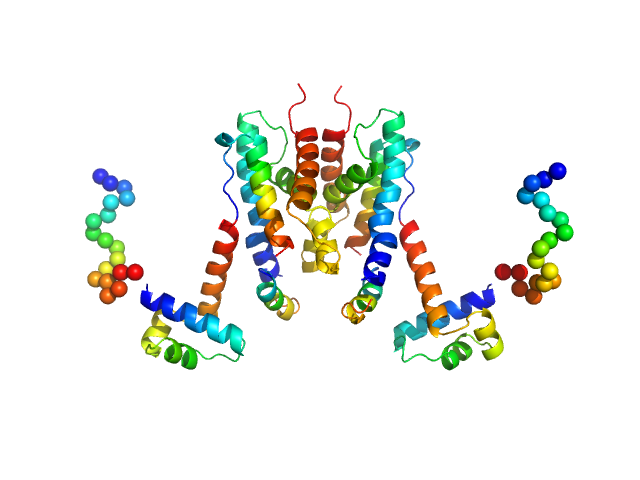
|
| Sample: |
TetR/AcrR family transcriptional regulator dimer, 48 kDa Bradyrhizobium diazoefficiens protein
|
| Buffer: |
20 mM Na2HPO4, 50 mM NaCl, 50 mM imidazole, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Sep 23
|
The Induction Mechanism of the Flavonoid-Responsive Regulator FrrA.
FEBS J (2021)
Werner N, Werten S, Hoppen J, Palm GJ, Göttfert M, Hinrichs W
|
| RgGuinier |
3.4 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
93 |
nm3 |
|
|
|
|
|
|
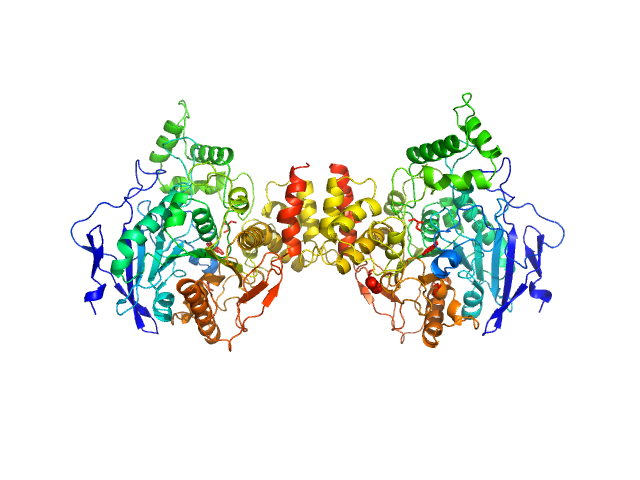
|
| Sample: |
Acetylcholinesterase dimer, 120 kDa Homo sapiens protein
Acetylcholinesterase monomer, 60 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris/HCl, 100 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2015 May 29
|
Covalent inhibition of hAChE by organophosphates causes homodimer dissociation through long-range allosteric effects.
J Biol Chem :101007 (2021)
Blumenthal DK, Cheng X, Fajer M, Ho KY, Rohrer J, Gerlits O, Taylor P, Juneja P, Kovalevsky A, Radić Z
|
| RgGuinier |
3.9 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
162 |
nm3 |
|
|
|
|
|
|
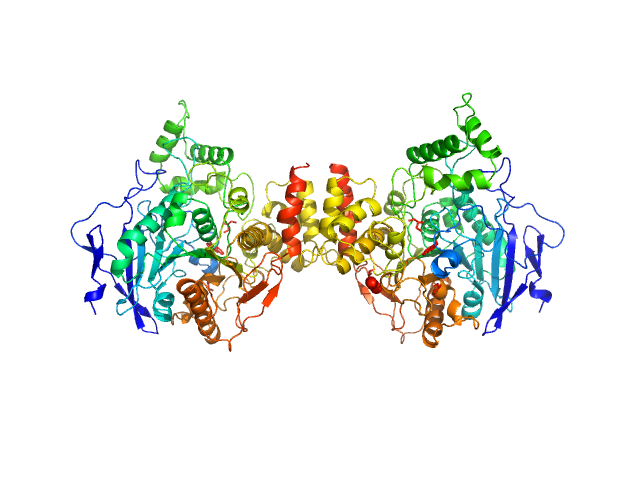
|
| Sample: |
Acetylcholinesterase dimer, 120 kDa Homo sapiens protein
Acetylcholinesterase monomer, 60 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris/HCl, 100 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2015 May 29
|
Covalent inhibition of hAChE by organophosphates causes homodimer dissociation through long-range allosteric effects.
J Biol Chem :101007 (2021)
Blumenthal DK, Cheng X, Fajer M, Ho KY, Rohrer J, Gerlits O, Taylor P, Juneja P, Kovalevsky A, Radić Z
|
| RgGuinier |
3.4 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
142 |
nm3 |
|
|
|
|
|
|
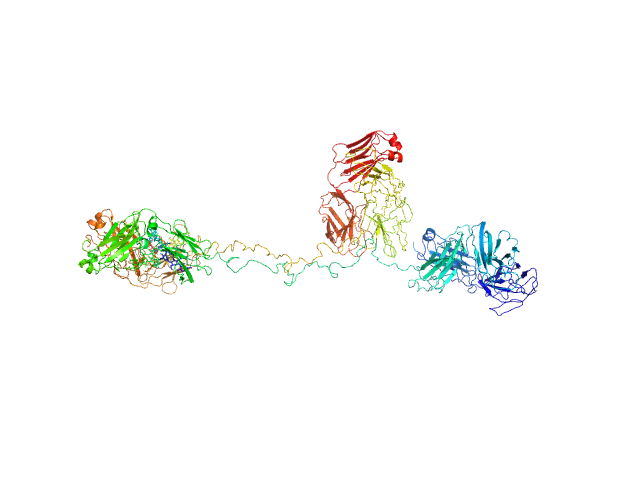
|
| Sample: |
Immunoglobulin G subclass 3 monomer, 155 kDa Homo sapiens protein
|
| Buffer: |
20 mM L-histidine, 138 mM NaCl, and 2.6 mM KCl buffer, pH: 6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Jul 29
|
Solution structures of human myeloma IgG3 antibody reveal extended Fab and Fc regions relative to the other IgG subclasses.
J Biol Chem :100995 (2021)
Spiteri VA, Goodall M, Doutch J, Rambo RP, Gor J, Perkins SJ
|
| RgGuinier |
7.0 |
nm |
| Dmax |
22.9 |
nm |
| VolumePorod |
267 |
nm3 |
|
|
|
|
|
|
|
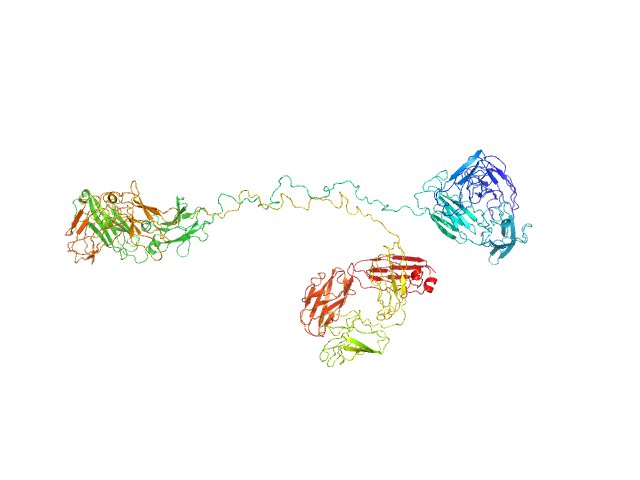
| Sample: |
Immunoglobulin G subclass 3 monomer, 155 kDa Homo sapiens protein
|
| Buffer: |
20 mM L-histidine, 138 mM NaCl, and 2.6 mM KCl buffer, pH: 6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Jul 29
|
Solution structures of human myeloma IgG3 antibody reveal extended Fab and Fc regions relative to the other IgG subclasses.
J Biol Chem :100995 (2021)
Spiteri VA, Goodall M, Doutch J, Rambo RP, Gor J, Perkins SJ
|
| RgGuinier |
7.1 |
nm |
| Dmax |
27.7 |
nm |
| VolumePorod |
294 |
nm3 |
|
|
|
|
|
|
|
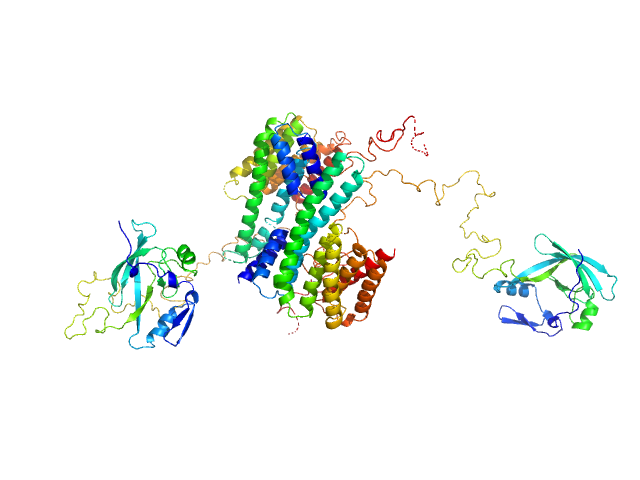
| Sample: |
14-3-3 protein zeta/delta dimer, 53 kDa Homo sapiens protein
Ataxin-1 AXH-C dimer, 55 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Oct 13
|
A structural study of the cytoplasmic chaperone effect of 14-3-3 proteins on Ataxin-1.
J Mol Biol :167174 (2021)
Leysen S, Jane Burnley R, Rodriguez E, Milroy LG, Soini L, Adamski CJ, Nitschke L, Davis R, Obsil T, Brunsveld L, Crabbe T, Yahya Zoghbi H, Ottmann C, Martin Davis J
|
| RgGuinier |
4.8 |
nm |
| Dmax |
19.8 |
nm |
|
|
|
|
|
|
|
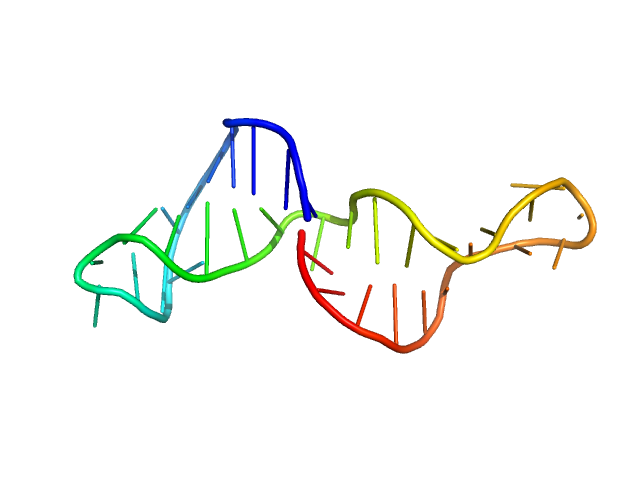
| Sample: |
Lung adenocarcinoma aptamer, truncated version monomer, 11 kDa Artificially synthesized DNA
|
| Buffer: |
Phosphate-buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Oct 28
|
The Role of Small-Angle X-Ray Scattering and Molecular Simulations in 3D Structure Elucidation of a DNA Aptamer Against Lung Cancer
Molecular Therapy - Nucleic Acids (2021)
Morozov D, Mironov V, Moryachkov R, Shchugoreva I, Artyushenko P, Zamay G, Kolovskaya O, Zamay T, Krat A, Molodenskiy D, Zabluda V, Veprintsev D, Sokolov A, Zukov R, Berezovski M, Tomilin F, Fedorov D, Alexeev Y, Kichkailo A
|
| RgGuinier |
2.1 |
nm |
| Dmax |
6.7 |
nm |
| VolumePorod |
16 |
nm3 |
|
|
|
|
|
|
|
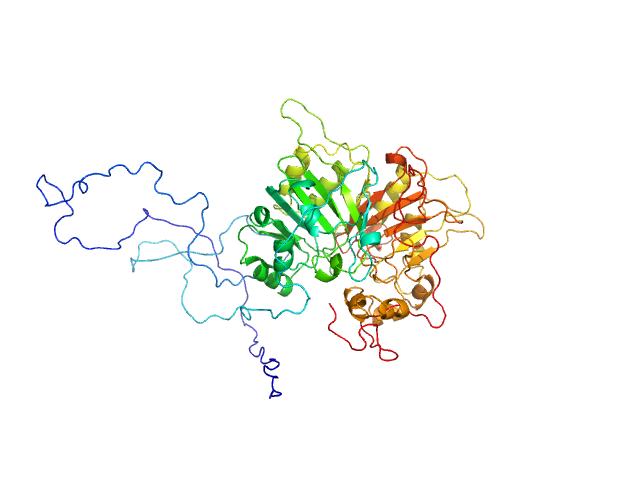
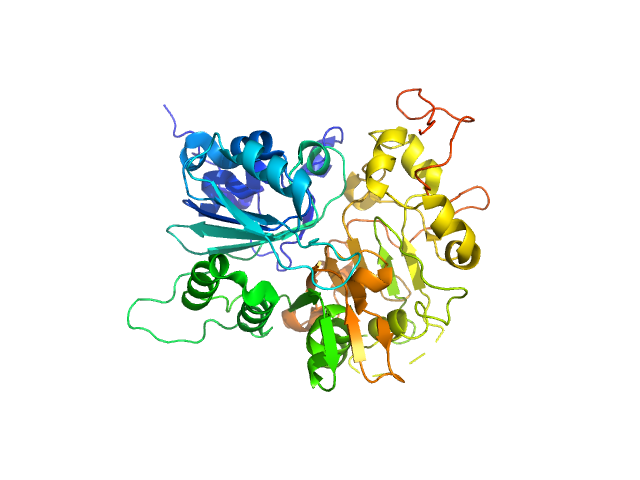
| Sample: |
Tyrosyl-DNA phosphodiesterase 1 monomer, 71 kDa Homo sapiens protein
|
| Buffer: |
200 mM NaCl, 20 mM Tris-HCl, pH 7.5, 2% glycerol,, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Jun 26
|
Direct interaction of DNA repair protein tyrosyl DNA phosphodiesterase 1 and the DNA ligase III catalytic domain is regulated by phosphorylation of its flexible N-terminus.
J Biol Chem :100921 (2021)
Rashid I, Hammel M, Sverzhinsky A, Tsai MS, Pascal JM, Tainer JA, Tomkinson AE
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.7 |
nm |
| VolumePorod |
143 |
nm3 |
|
|
|
|
|
|
|
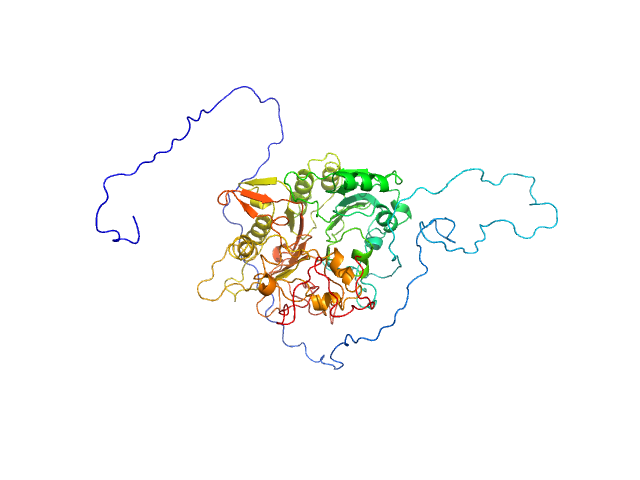
| Sample: |
Tyrosyl-DNA phosphodiesterase 1 (149-608) monomer, 53 kDa Homo sapiens protein
|
| Buffer: |
200 mM NaCl, 20 mM Tris-HCl, pH 7.5, 2% glycerol,, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Sep 18
|
Direct interaction of DNA repair protein tyrosyl DNA phosphodiesterase 1 and the DNA ligase III catalytic domain is regulated by phosphorylation of its flexible N-terminus.
J Biol Chem :100921 (2021)
Rashid I, Hammel M, Sverzhinsky A, Tsai MS, Pascal JM, Tainer JA, Tomkinson AE
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
80 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Tyrosyl-DNA phosphodiesterase 1 (dephosphorylated) monomer, 71 kDa Homo sapiens protein
|
| Buffer: |
200 mM NaCl, 20 mM Tris-HCl, pH 7.5, 2% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Jun 26
|
Direct interaction of DNA repair protein tyrosyl DNA phosphodiesterase 1 and the DNA ligase III catalytic domain is regulated by phosphorylation of its flexible N-terminus.
J Biol Chem :100921 (2021)
Rashid I, Hammel M, Sverzhinsky A, Tsai MS, Pascal JM, Tainer JA, Tomkinson AE
|
| RgGuinier |
3.2 |
nm |
| Dmax |
16.5 |
nm |
| VolumePorod |
141 |
nm3 |
|
|