|
|
|
|
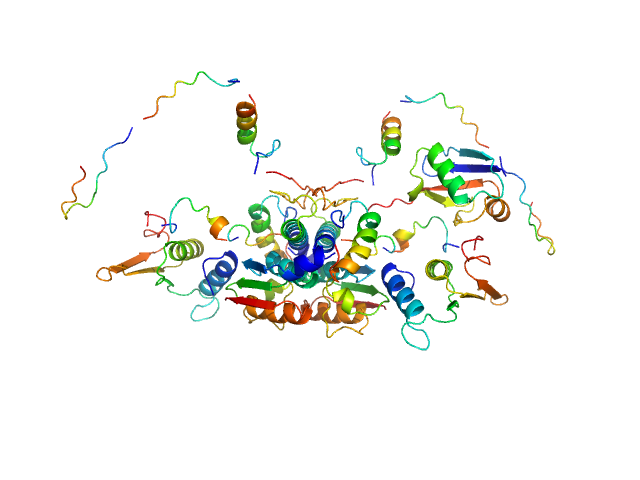
|
| Sample: |
Ubiquitin-like modifier-activating enzyme 5 dimer, 68 kDa Homo sapiens protein
Ubiquitin fold modifer 1 monomer, 9 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 50 mM NaCl, 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Oct 29
|
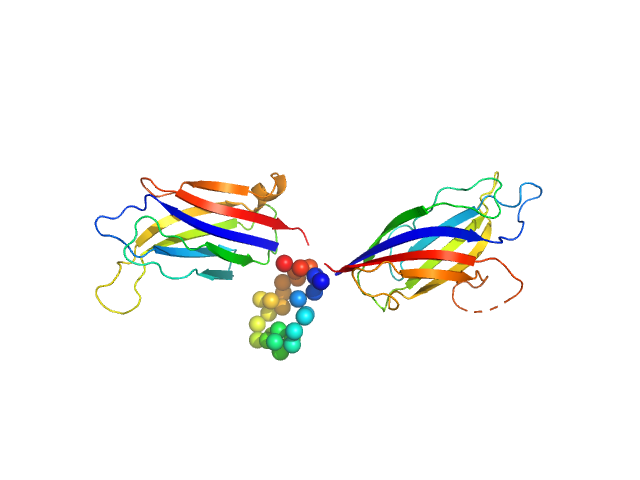
Structure and dynamics of UBA5-UFM1 complex formation showing new insights in the UBA5 activation mechanism
Journal of Structural Biology :107796 (2021)
Fuchs S, Kikhney A, Schubert R, Kaiser C, Liebau E, Svergun D, Betzel C, Perbandt M
|
| RgGuinier |
3.2 |
nm |
| Dmax |
14.5 |
nm |
| VolumePorod |
111 |
nm3 |
|
|
|
|
|
|
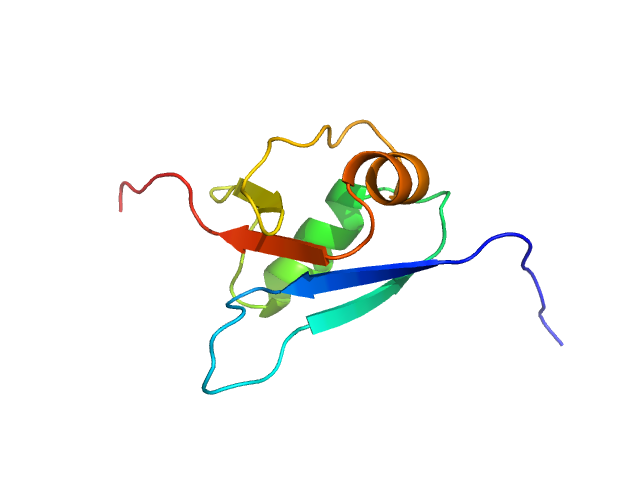
|
| Sample: |
Ubiquitin fold modifer 1 monomer, 9 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Jun 13
|
Structure and dynamics of UBA5-UFM1 complex formation showing new insights in the UBA5 activation mechanism
Journal of Structural Biology :107796 (2021)
Fuchs S, Kikhney A, Schubert R, Kaiser C, Liebau E, Svergun D, Betzel C, Perbandt M
|
| RgGuinier |
1.5 |
nm |
| Dmax |
5.1 |
nm |
| VolumePorod |
16 |
nm3 |
|
|
|
|
|
|
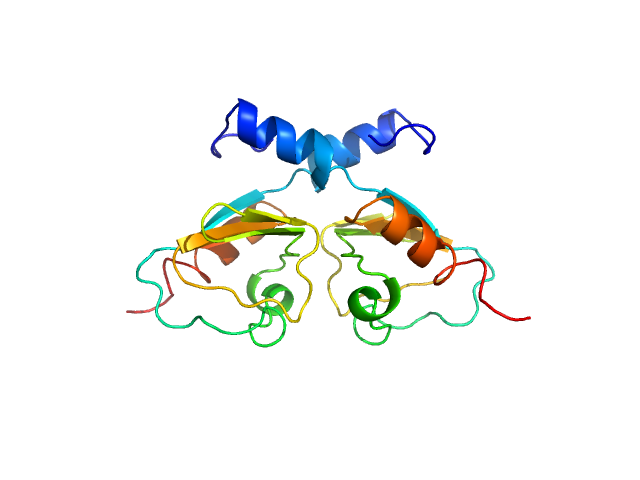
|
| Sample: |
Transcriptional repressor BusR RCK_C domain dimer, 22 kDa Streptococcus agalactiae serotype … protein
|
| Buffer: |
100 mM NaCl, 30mM Hepes, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Jul 2
|
BusR senses bipartite DNA binding motifs by a unique molecular ruler architecture.
Nucleic Acids Res (2021)
Bandera AM, Bartho J, Lammens K, Drexler DJ, Kleinschwärzer J, Hopfner KP, Witte G
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.4 |
nm |
| VolumePorod |
44 |
nm3 |
|
|
|
|
|
|
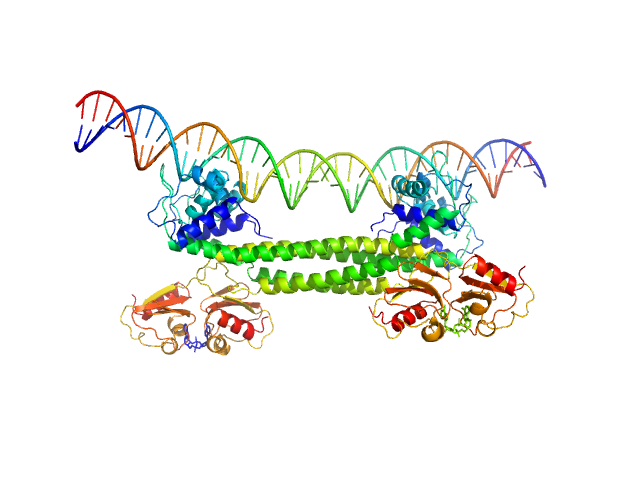
|
| Sample: |
Transcriptional repressor BusR tetramer, 95 kDa Streptococcus agalactiae protein
BusR Recognition sequence monomer, 28 kDa synthetic construct DNA
|
| Buffer: |
20mM HEPES, pH6.5, 100mM NaCl, 3% glycerol (v/v), pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Jul 2
|
BusR senses bipartite DNA binding motifs by a unique molecular ruler architecture.
Nucleic Acids Res (2021)
Bandera AM, Bartho J, Lammens K, Drexler DJ, Kleinschwärzer J, Hopfner KP, Witte G
|
| RgGuinier |
4.3 |
nm |
| Dmax |
14.2 |
nm |
| VolumePorod |
210 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Synaptotagmin-1 monomer, 33 kDa Arabidopsis thaliana protein
|
| Buffer: |
50 mM Tris, 50 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2020 Jun 11
|
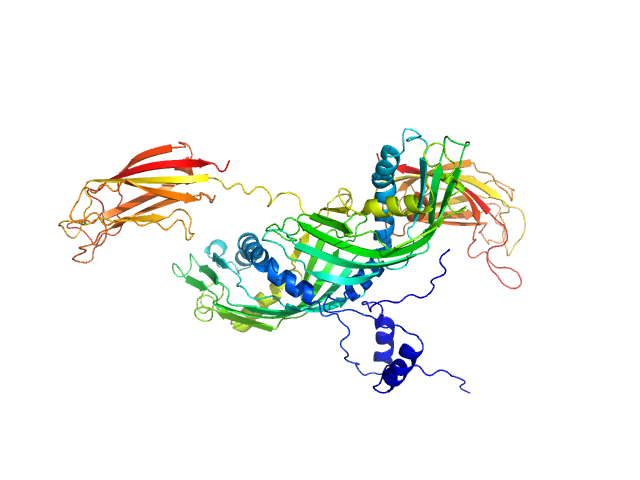
The structure and flexibility analysis of the Arabidopsis
synaptotagmin 1 reveal the basis of its regulation at membrane contact sites
Life Science Alliance 4(10):e202101152 (2021)
Benavente J, Siliqi D, Infantes L, Lagartera L, Mills A, Gago F, Ruiz-López N, Botella M, Sánchez-Barrena M, Albert A
|
| RgGuinier |
3.1 |
nm |
| Dmax |
12.1 |
nm |
| VolumePorod |
59 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Synaptotagmin-1 (SYT1-SMP2C2A) monomer, 72 kDa Escherichia coli protein
|
| Buffer: |
20 mM Tris, 100 mM NaCl, 5% glycerol, 2 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2020 Jun 11
|
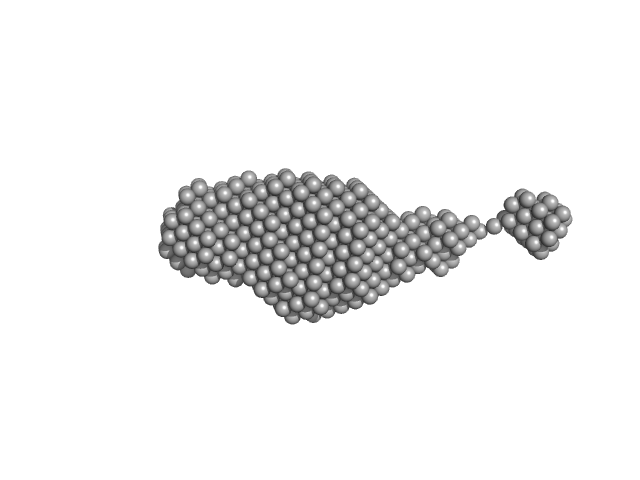
The structure and flexibility analysis of the Arabidopsis
synaptotagmin 1 reveal the basis of its regulation at membrane contact sites
Life Science Alliance 4(10):e202101152 (2021)
Benavente J, Siliqi D, Infantes L, Lagartera L, Mills A, Gago F, Ruiz-López N, Botella M, Sánchez-Barrena M, Albert A
|
| RgGuinier |
4.2 |
nm |
| Dmax |
17.8 |
nm |
| VolumePorod |
138 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Synaptotagmin-1 monomer, 33 kDa Arabidopsis thaliana protein
|
| Buffer: |
50 mM Tris, 50 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2021 Feb 16
|
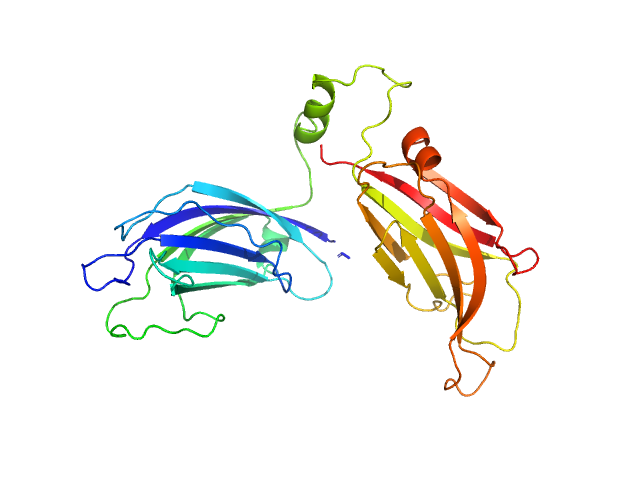
The structure and flexibility analysis of the Arabidopsis
synaptotagmin 1 reveal the basis of its regulation at membrane contact sites
Life Science Alliance 4(10):e202101152 (2021)
Benavente J, Siliqi D, Infantes L, Lagartera L, Mills A, Gago F, Ruiz-López N, Botella M, Sánchez-Barrena M, Albert A
|
| RgGuinier |
2.8 |
nm |
| Dmax |
12.6 |
nm |
| VolumePorod |
50 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Synaptotagmin-1 monomer, 33 kDa Arabidopsis thaliana protein
|
| Buffer: |
50 mM Tris, 50 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2021 Feb 16
|
The structure and flexibility analysis of the Arabidopsis
synaptotagmin 1 reveal the basis of its regulation at membrane contact sites
Life Science Alliance 4(10):e202101152 (2021)
Benavente J, Siliqi D, Infantes L, Lagartera L, Mills A, Gago F, Ruiz-López N, Botella M, Sánchez-Barrena M, Albert A
|
| RgGuinier |
2.9 |
nm |
| Dmax |
13.8 |
nm |
| VolumePorod |
54 |
nm3 |
|
|
|
|
|
|
|
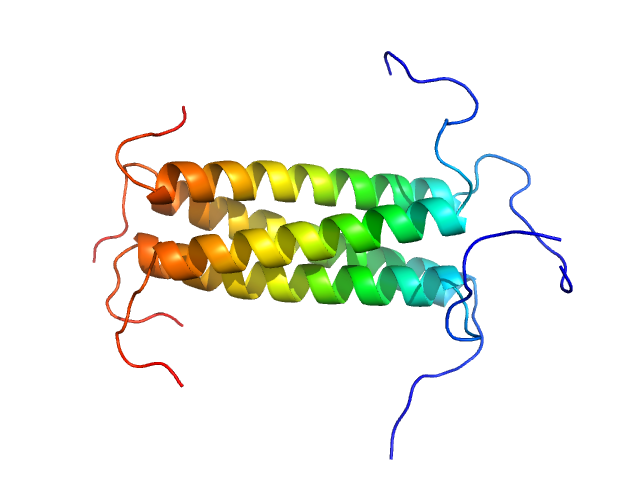
| Sample: |
Phosphoprotein tetramer, 18 kDa Respiratory syncytial virus protein
|
| Buffer: |
20 mM Na phosphate 100 mM NaCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Jun 21
|
A Structural and Dynamic Analysis of the Partially Disordered Polymerase-Binding Domain in RSV Phosphoprotein
Biomolecules 11(8):1225 (2021)
Cardone C, Caseau C, Bardiaux B, Thureaux A, Galloux M, Bajorek M, Eléouët J, Litaudon M, Bontems F, Sizun C
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.6 |
nm |
| VolumePorod |
33 |
nm3 |
|
|
|
|
|
|
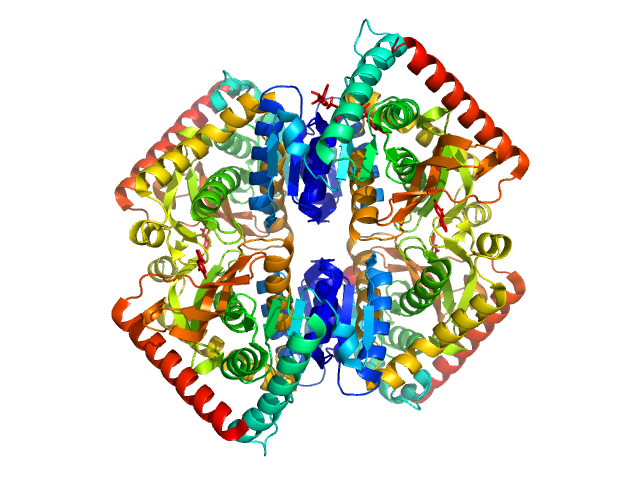
|
| Sample: |
L-lactate dehydrogenase tetramer, 141 kDa Plasmodium falciparum protein
|
| Buffer: |
100 mM Na-phosphate buffer, 400 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at Xenocs Xeuss 2.0 with MetalJet, Department of Macromolecular Physics, Faculty of Physics, Adam Mickiewicz University on 2019 Jul 3
|
A fragment-based approach identifies an allosteric pocket that impacts malate dehydrogenase activity
Communications Biology 4(1) (2021)
Reyes Romero A, Lunev S, Popowicz G, Calderone V, Gentili M, Sattler M, Plewka J, Taube M, Kozak M, Holak T, Dömling A, Groves M
|
| RgGuinier |
3.4 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
211 |
nm3 |
|
|