|
|
|
|

|
| Sample: |
DNA ligase A monomer, 38 kDa Mycobacterium tuberculosis protein
|
| Buffer: |
50 mM Tris-HCl, 200 mM NaCl, 2 mM β-mercaptoethanol, pH: 8 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR-Central Drug Research Institute on 2018 Sep 27
|
Salt bridges at the subdomain interfaces of the adenylation domain and active-site residues of Mycobacterium tuberculosis
NAD +
-dependent DNA ligase A (MtbLigA) are important for the initial steps of nick-sealing activity
Acta Crystallographica Section D Structural Biology 77(6) (2021)
Afsar M, Shukla A, Kumar N, Ramachandran R
|
| RgGuinier |
2.4 |
nm |
| Dmax |
8.8 |
nm |
| VolumePorod |
69 |
nm3 |
|
|
|
|
|
|
|
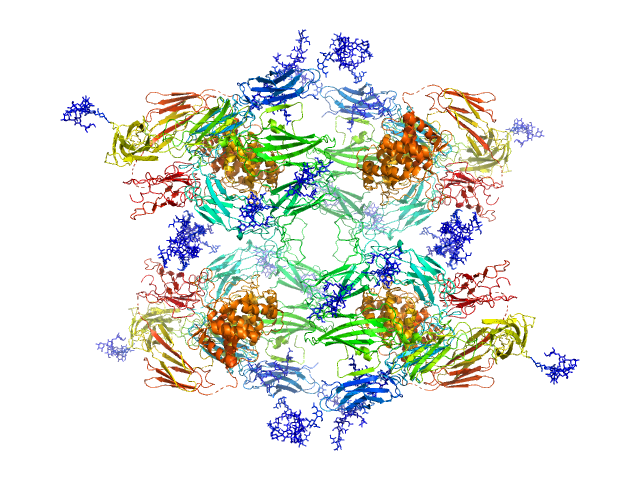
| Sample: |
Alpha-2-macroglobulin tetramer, 643 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at Bruker Nanostar w Excillum source, Department of Chemistry, iNANO building, Aarhus Uinversity on 2020 Jan 22
|
Structural Investigations of Human A2M Identify a Hollow Native Conformation That Underlies Its Distinctive Protease-Trapping Mechanism.
Mol Cell Proteomics 20:100090 (2021)
Harwood SL, Lyngsø J, Zarantonello A, Kjøge K, Nielsen PK, Andersen GR, Pedersen JS, Enghild JJ
|
| RgGuinier |
7.7 |
nm |
| Dmax |
22.7 |
nm |
|
|
|
|
|
|
|
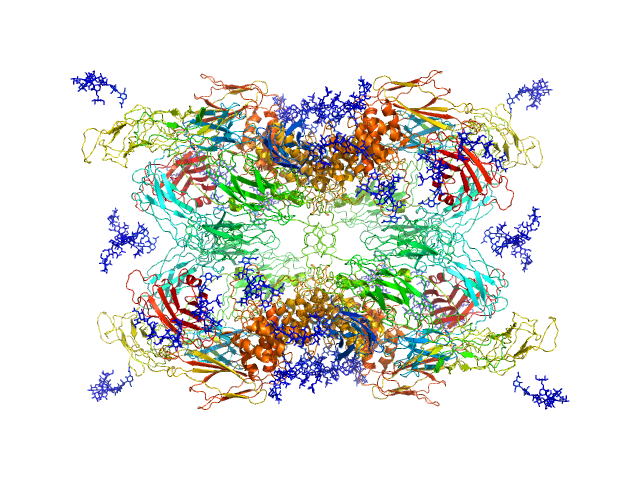
| Sample: |
Alpha-2-macroglobulin tetramer, 643 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at Bruker Nanostar w Excillum source, Department of Chemistry, iNANO building, Aarhus Uinversity on 2020 Jan 22
|
Structural Investigations of Human A2M Identify a Hollow Native Conformation That Underlies Its Distinctive Protease-Trapping Mechanism.
Mol Cell Proteomics 20:100090 (2021)
Harwood SL, Lyngsø J, Zarantonello A, Kjøge K, Nielsen PK, Andersen GR, Pedersen JS, Enghild JJ
|
| RgGuinier |
6.7 |
nm |
| Dmax |
19.7 |
nm |
|
|
|
|
|
|
|
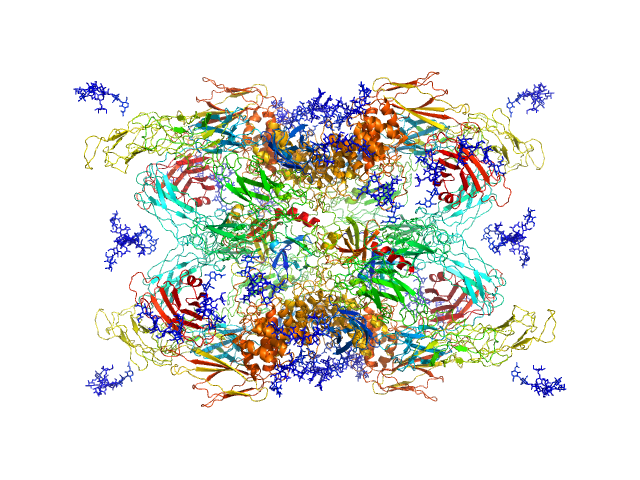
| Sample: |
Cationic trypsin dimer, 52 kDa Bos taurus protein
Alpha-2-macroglobulin tetramer, 643 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at Bruker Nanostar w Excillum source, Department of Chemistry, iNANO building, Aarhus Uinversity on 2020 Jan 15
|
Structural Investigations of Human A2M Identify a Hollow Native Conformation That Underlies Its Distinctive Protease-Trapping Mechanism.
Mol Cell Proteomics 20:100090 (2021)
Harwood SL, Lyngsø J, Zarantonello A, Kjøge K, Nielsen PK, Andersen GR, Pedersen JS, Enghild JJ
|
| RgGuinier |
6.6 |
nm |
| Dmax |
19.7 |
nm |
|
|
|
|
|
|
|
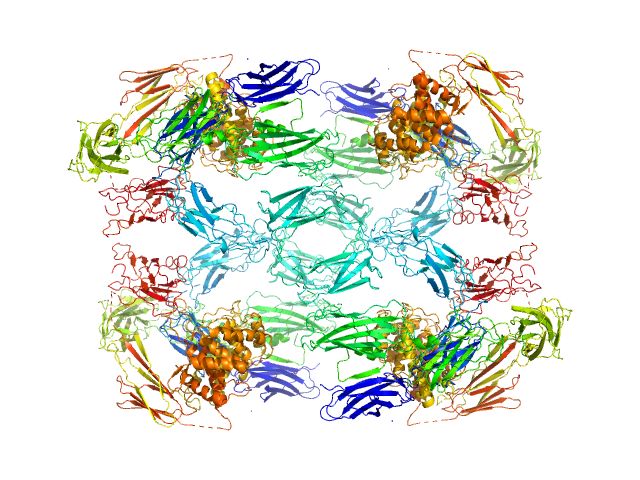
| Sample: |
Alpha-2-macroglobulin tetramer, 643 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at Bruker Nanostar w Excillum source, Department of Chemistry, iNANO building, Aarhus Uinversity on 2019 Apr 12
|
Structural Investigations of Human A2M Identify a Hollow Native Conformation That Underlies Its Distinctive Protease-Trapping Mechanism.
Mol Cell Proteomics 20:100090 (2021)
Harwood SL, Lyngsø J, Zarantonello A, Kjøge K, Nielsen PK, Andersen GR, Pedersen JS, Enghild JJ
|
| RgGuinier |
7.6 |
nm |
| Dmax |
20.2 |
nm |
|
|
|
|
|
|
|
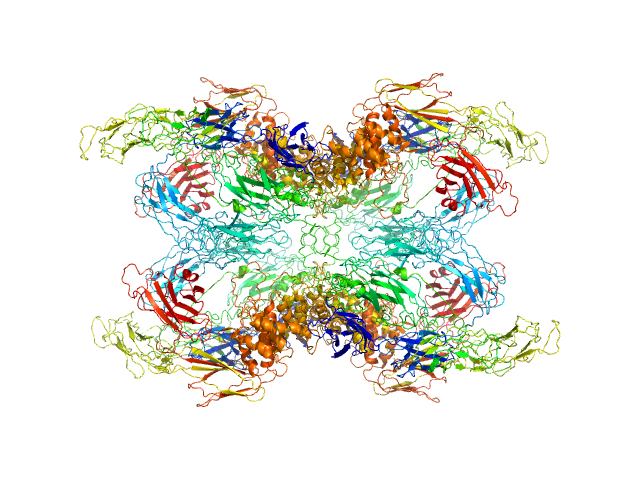
| Sample: |
Alpha-2-macroglobulin tetramer, 643 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at Bruker Nanostar w Excillum source, Department of Chemistry, iNANO building, Aarhus Uinversity on 2019 Apr 13
|
Structural Investigations of Human A2M Identify a Hollow Native Conformation That Underlies Its Distinctive Protease-Trapping Mechanism.
Mol Cell Proteomics 20:100090 (2021)
Harwood SL, Lyngsø J, Zarantonello A, Kjøge K, Nielsen PK, Andersen GR, Pedersen JS, Enghild JJ
|
| RgGuinier |
6.6 |
nm |
| Dmax |
19.7 |
nm |
|
|
|
|
|
|
|
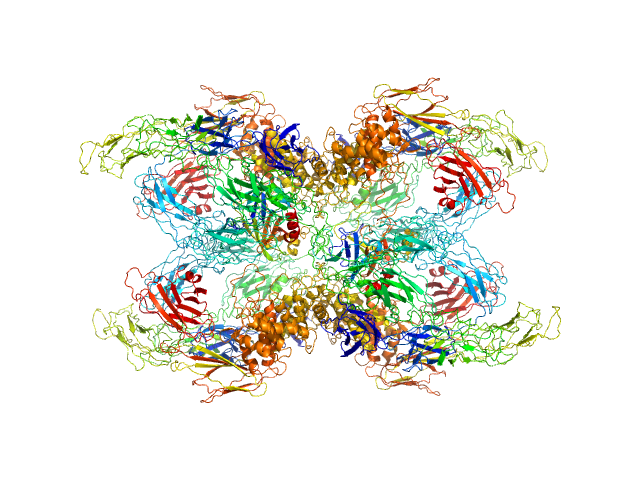
| Sample: |
Cationic trypsin dimer, 52 kDa Bos taurus protein
Alpha-2-macroglobulin tetramer, 643 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at Bruker Nanostar w Excillum source, Department of Chemistry, iNANO building, Aarhus Uinversity on 2020 Jan 22
|
Structural Investigations of Human A2M Identify a Hollow Native Conformation That Underlies Its Distinctive Protease-Trapping Mechanism.
Mol Cell Proteomics 20:100090 (2021)
Harwood SL, Lyngsø J, Zarantonello A, Kjøge K, Nielsen PK, Andersen GR, Pedersen JS, Enghild JJ
|
| RgGuinier |
6.6 |
nm |
| Dmax |
20.7 |
nm |
|
|
|
|
|
|
|
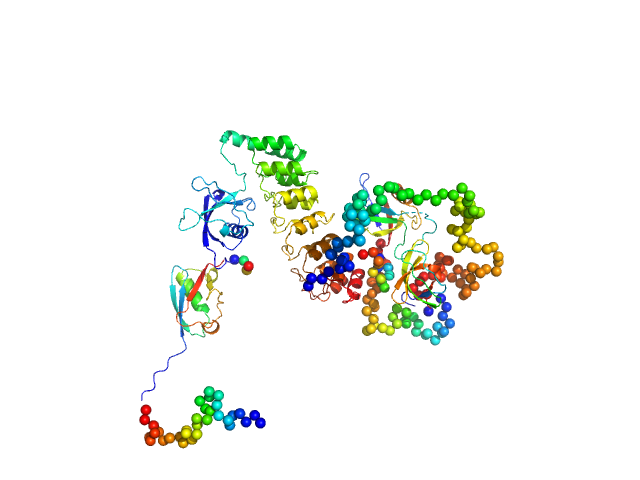
| Sample: |
SH3 and multiple ankyrin repeat domains protein 3 monomer, 88 kDa Rattus norvegicus protein
|
| Buffer: |
100mM NaH2PO4, 100mM NaCl, 0.5mM DTT,, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Jun 3
|
Autism associated SHANK3 missense point mutations impact conformational fluctuations and protein turnover at synapses.
Elife 10 (2021)
Bucher M, Niebling S, Han Y, Molodenskiy D, Nia FH, Kreienkamp HJ, Svergun D, Kim E, Kostyukova AS, Kreutz MR, Mikhaylova M
|
| RgGuinier |
4.1 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
170 |
nm3 |
|
|
|
|
|
|
|
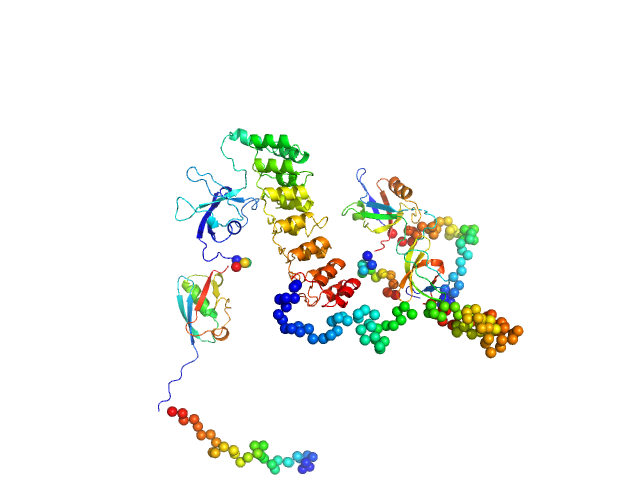
| Sample: |
SH3 and multiple ankyrin repeat domains protein 3 monomer, 88 kDa Rattus norvegicus protein
|
| Buffer: |
100mM NaH2PO4, 100mM NaCl, 0.5mM DTT,, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Jun 3
|
Autism associated SHANK3 missense point mutations impact conformational fluctuations and protein turnover at synapses.
Elife 10 (2021)
Bucher M, Niebling S, Han Y, Molodenskiy D, Nia FH, Kreienkamp HJ, Svergun D, Kim E, Kostyukova AS, Kreutz MR, Mikhaylova M
|
| RgGuinier |
4.1 |
nm |
| Dmax |
14.8 |
nm |
| VolumePorod |
224 |
nm3 |
|
|
|
|
|
|
|
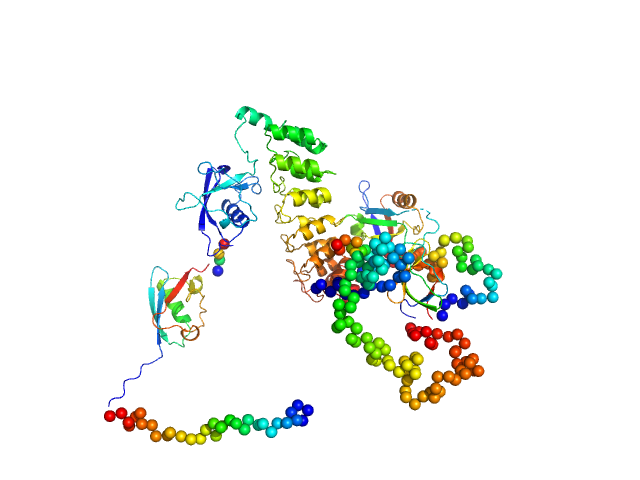
| Sample: |
SH3 and multiple ankyrin repeat domains protein 3 monomer, 87 kDa Rattus norvegicus protein
|
| Buffer: |
100mM NaH2PO4, 100mM NaCl, 0.5mM DTT,, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Jun 3
|
Autism associated SHANK3 missense point mutations impact conformational fluctuations and protein turnover at synapses.
Elife 10 (2021)
Bucher M, Niebling S, Han Y, Molodenskiy D, Nia FH, Kreienkamp HJ, Svergun D, Kim E, Kostyukova AS, Kreutz MR, Mikhaylova M
|
| RgGuinier |
4.1 |
nm |
| Dmax |
13.8 |
nm |
| VolumePorod |
175 |
nm3 |
|
|