|
|
|
|
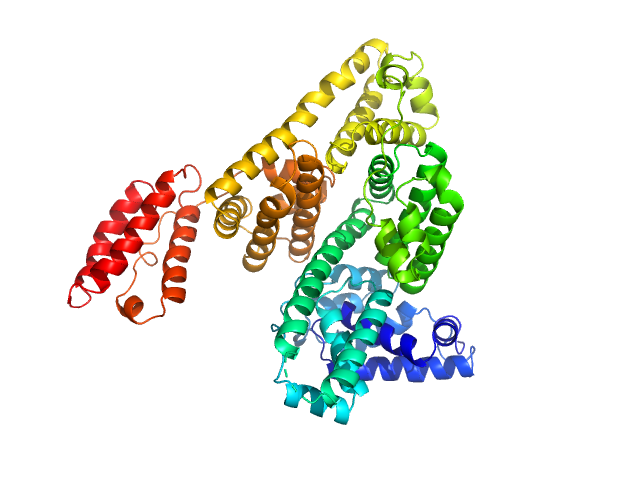
|
| Sample: |
Albumin monomer, 69 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM KCl, 2% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Dec 1
|
Albumin in patients with liver disease shows an altered conformation.
Commun Biol 4(1):731 (2021)
Paar M, Fengler VH, Rosenberg DJ, Krebs A, Stauber RE, Oettl K, Hammel M
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.4 |
nm |
|
|
|
|
|
|
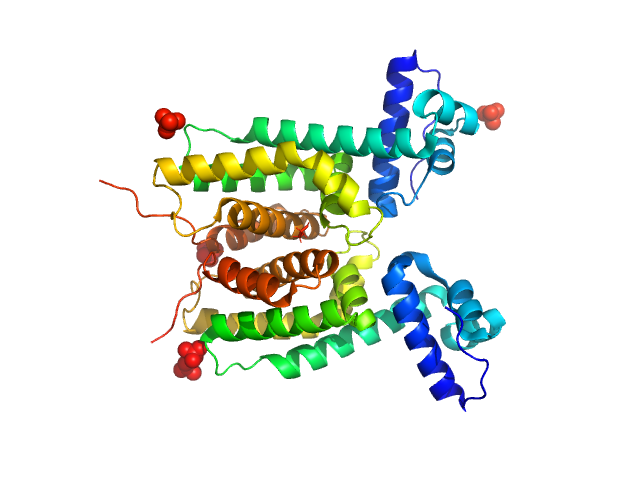
|
| Sample: |
LuxR family transcriptional regulator, 52 kDa Vibrio vulnificus protein
|
| Buffer: |
200 mM NaCl, 25 mM Tris, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Aug 8
|
The DNA binding domain of the Vibrio vulnificus SmcR transcription factor is flexible and binds diverse DNA sequences.
Nucleic Acids Res 49(10):5967-5984 (2021)
Newman JD, Russell MM, Fan L, Wang YX, Gonzalez-Gutierrez G, van Kessel JC
|
| RgGuinier |
3.1 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
67 |
nm3 |
|
|
|
|
|
|
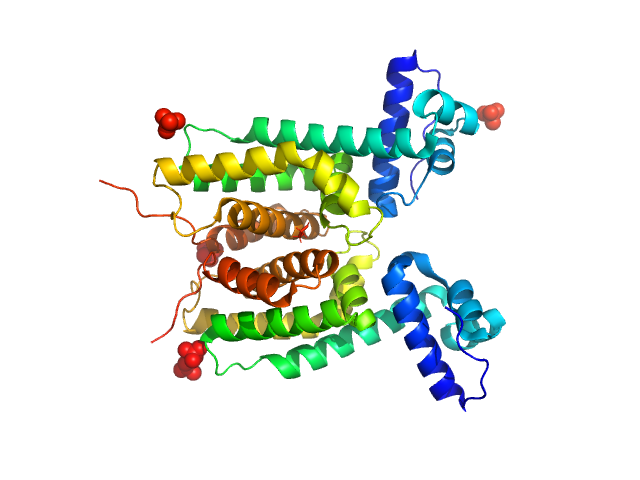
|
| Sample: |
LuxR family transcriptional regulator, N55I, 52 kDa Vibrio vulnificus protein
|
| Buffer: |
200 mM NaCl, 25 mM Tris, 1 mM DTT, 0.5 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Dec 11
|
The DNA binding domain of the Vibrio vulnificus SmcR transcription factor is flexible and binds diverse DNA sequences.
Nucleic Acids Res 49(10):5967-5984 (2021)
Newman JD, Russell MM, Fan L, Wang YX, Gonzalez-Gutierrez G, van Kessel JC
|
| RgGuinier |
3.0 |
nm |
| Dmax |
10.1 |
nm |
| VolumePorod |
45 |
nm3 |
|
|
|
|
|
|
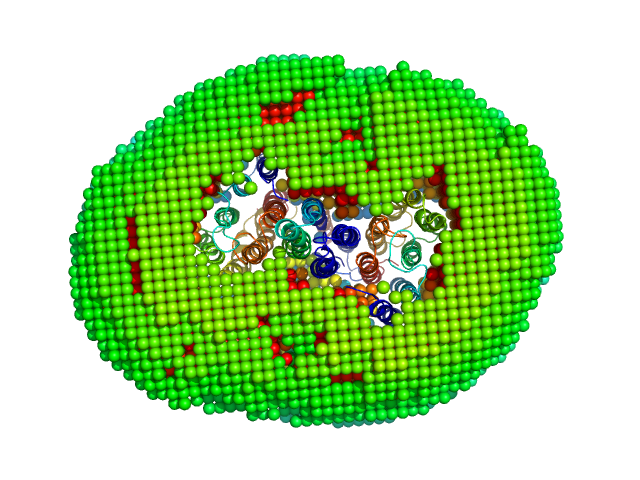
|
| Sample: |
Sensory rhodopsin II from Natronbacterium pharaonis dimer, 53 kDa Natronomonas pharaonis protein
Truncated sensory rhodopsin II transducer containing transmembrane and HAMP1 domains dimer, 30 kDa Natronomonas pharaonis protein
|
| Buffer: |
150 mM NaCl, 25 Na/K-Pi, 1 mM EDTA, 0.05 % DDM, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Dec 13
|
Molecular model of a sensor of two-component signaling system
Scientific Reports 11(1) (2021)
Ryzhykau Y, Orekhov P, Rulev M, Vlasov A, Melnikov I, Volkov D, Nikolaev M, Zabelskii D, Murugova T, Chupin V, Rogachev A, Gruzinov A, Svergun D, Brennich M, Gushchin I, Soler-Lopez M, Bothe A, Büldt G, Leonard G, Engelhard M, Kuklin A, Gordeliy V
|
| RgGuinier |
4.8 |
nm |
| Dmax |
14.0 |
nm |
|
|
|
|
|
|

|
| Sample: |
Sensory rhodopsin II from Natronbacterium pharaonis dimer, 53 kDa Natronomonas pharaonis protein
Sensory rhodopsin II transducer from Natronomonas pharaonis dimer, 116 kDa Natronomonas pharaonis protein
|
| Buffer: |
150 mM NaCl, 25 mM Na/Na-Pi, 1.0 mM EDTA, 0.05% DDM, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Jul 9
|
Molecular model of a sensor of two-component signaling system
Scientific Reports 11(1) (2021)
Ryzhykau Y, Orekhov P, Rulev M, Vlasov A, Melnikov I, Volkov D, Nikolaev M, Zabelskii D, Murugova T, Chupin V, Rogachev A, Gruzinov A, Svergun D, Brennich M, Gushchin I, Soler-Lopez M, Bothe A, Büldt G, Leonard G, Engelhard M, Kuklin A, Gordeliy V
|
| RgGuinier |
9.0 |
nm |
| Dmax |
38.9 |
nm |
|
|
|
|
|
|

|
| Sample: |
Sensory rhodopsin II from Natronbacterium pharaonis dimer, 53 kDa Natronomonas pharaonis protein
Sensory rhodopsin II transducer from Natronomonas pharaonis dimer, 116 kDa Natronomonas pharaonis protein
|
| Buffer: |
150 mM NaCl, 25 mM Na/Na-Pi, 1.0 mM EDTA, 0.05% DDM (D2O buffer), pH: 8 |
| Experiment: |
SANS
data collected at YuMO SANS TOF spectrometer, IBR-2, Frank Laboratory of Neutron Physics, Joint Institute for Nuclear Research on 2019 Jan 25
|
Molecular model of a sensor of two-component signaling system
Scientific Reports 11(1) (2021)
Ryzhykau Y, Orekhov P, Rulev M, Vlasov A, Melnikov I, Volkov D, Nikolaev M, Zabelskii D, Murugova T, Chupin V, Rogachev A, Gruzinov A, Svergun D, Brennich M, Gushchin I, Soler-Lopez M, Bothe A, Büldt G, Leonard G, Engelhard M, Kuklin A, Gordeliy V
|
| RgGuinier |
7.1 |
nm |
| Dmax |
35.0 |
nm |
|
|
|
|
|
|

|
| Sample: |
Sensory rhodopsin II from Natronbacterium pharaonis dimer, 53 kDa Natronomonas pharaonis protein
Sensory rhodopsin II transducer from Natronomonas pharaonis dimer, 116 kDa Natronomonas pharaonis protein
|
| Buffer: |
1400 mM NaCl, 49.4 mM Na/Na-Pi, 1.0 mM EDTA, 0.05% DDM (D2O buffer), pH: 8 |
| Experiment: |
SANS
data collected at YuMO SANS TOF spectrometer, IBR-2, Frank Laboratory of Neutron Physics, Joint Institute for Nuclear Research on 2019 Feb 10
|
Molecular model of a sensor of two-component signaling system
Scientific Reports 11(1) (2021)
Ryzhykau Y, Orekhov P, Rulev M, Vlasov A, Melnikov I, Volkov D, Nikolaev M, Zabelskii D, Murugova T, Chupin V, Rogachev A, Gruzinov A, Svergun D, Brennich M, Gushchin I, Soler-Lopez M, Bothe A, Büldt G, Leonard G, Engelhard M, Kuklin A, Gordeliy V
|
| RgGuinier |
9.9 |
nm |
| Dmax |
36.5 |
nm |
|
|
|
|
|
|

|
| Sample: |
Sensory rhodopsin II from Natronbacterium pharaonis dimer, 53 kDa Natronomonas pharaonis protein
Sensory rhodopsin II transducer from Natronomonas pharaonis dimer, 116 kDa Natronomonas pharaonis protein
|
| Buffer: |
2800 mM NaCl, 76.6 mM Na/Na-Pi, 1.0 mM EDTA, 0.05% DDM (D2O buffer), pH: 8 |
| Experiment: |
SANS
data collected at YuMO SANS TOF spectrometer, IBR-2, Frank Laboratory of Neutron Physics, Joint Institute for Nuclear Research on 2019 Feb 10
|
Molecular model of a sensor of two-component signaling system
Scientific Reports 11(1) (2021)
Ryzhykau Y, Orekhov P, Rulev M, Vlasov A, Melnikov I, Volkov D, Nikolaev M, Zabelskii D, Murugova T, Chupin V, Rogachev A, Gruzinov A, Svergun D, Brennich M, Gushchin I, Soler-Lopez M, Bothe A, Büldt G, Leonard G, Engelhard M, Kuklin A, Gordeliy V
|
| RgGuinier |
9.3 |
nm |
| Dmax |
37.5 |
nm |
|
|
|
|
|
|

|
| Sample: |
Sensory rhodopsin II from Natronbacterium pharaonis dimer, 53 kDa Natronomonas pharaonis protein
Sensory rhodopsin II transducer from Natronomonas pharaonis dimer, 116 kDa Natronomonas pharaonis protein
|
| Buffer: |
4000 mM NaCl, 100 mM Na/Na-Pi, 1.0 mM EDTA, 0.05% DDM (D2O buffer), pH: 8 |
| Experiment: |
SANS
data collected at YuMO SANS TOF spectrometer, IBR-2, Frank Laboratory of Neutron Physics, Joint Institute for Nuclear Research on 2019 Jan 25
|
Molecular model of a sensor of two-component signaling system
Scientific Reports 11(1) (2021)
Ryzhykau Y, Orekhov P, Rulev M, Vlasov A, Melnikov I, Volkov D, Nikolaev M, Zabelskii D, Murugova T, Chupin V, Rogachev A, Gruzinov A, Svergun D, Brennich M, Gushchin I, Soler-Lopez M, Bothe A, Büldt G, Leonard G, Engelhard M, Kuklin A, Gordeliy V
|
| RgGuinier |
8.9 |
nm |
| Dmax |
39.0 |
nm |
|
|
|
|
|
|
|
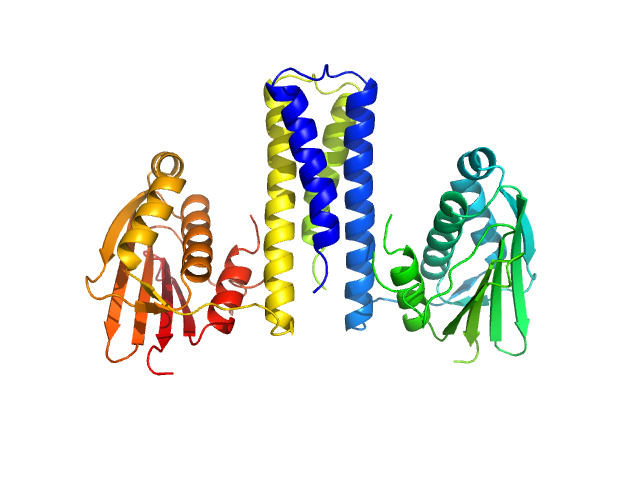
| Sample: |
Histidine kinase dimer, 51 kDa Acinetobacter baumannii protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2018 Apr 21
|
Proteolysis and multimerization regulate signaling along the two-component regulatory system AdeRS.
iScience 24(5):102476 (2021)
Ouyang Z, Zheng F, Zhu L, Felix J, Wu D, Wu K, Gutsche I, Wu Y, Hwang PM, She J, Wen Y
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.1 |
nm |
| VolumePorod |
85 |
nm3 |
|
|