|
|
|
|
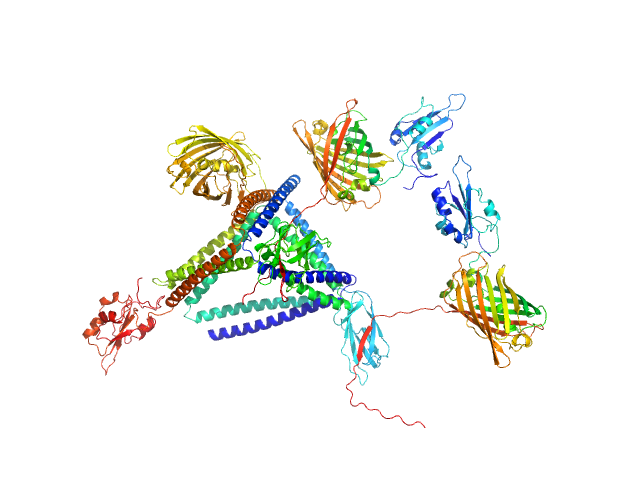
|
| Sample: |
TET12-2Sc-RFP-DCV monomer, 109 kDa synthetic construct protein
DCV-GFP-St monomer, 41 kDa synthetic construct protein
DCV-GFP-St monomer, 41 kDa synthetic construct protein
|
| Buffer: |
20 mM Tris, 150 mM NaCL, 3% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2024 Mar 18
|
Programmable Protein-DNA Composite Nanostructures: from Nanostructure Construction to Protein-Induced Micro-Scale Material Self-Assembly and Functionalization.
Small 21(30):e2502060 (2025)
Zhou W, Strmšek Ž, Snoj J, Škarabot M, Jerala R
|
| RgGuinier |
5.8 |
nm |
| Dmax |
17.3 |
nm |
| VolumePorod |
580 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
DCV-RFP-St monomer, 41 kDa synthetic construct protein
|
| Buffer: |
20 mM Tris, 150 mM NaCL, 3% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2024 Sep 24
|
Programmable Protein-DNA Composite Nanostructures: from Nanostructure Construction to Protein-Induced Micro-Scale Material Self-Assembly and Functionalization.
Small 21(30):e2502060 (2025)
Zhou W, Strmšek Ž, Snoj J, Škarabot M, Jerala R
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.2 |
nm |
| VolumePorod |
60 |
nm3 |
|
|
|
|
|
|
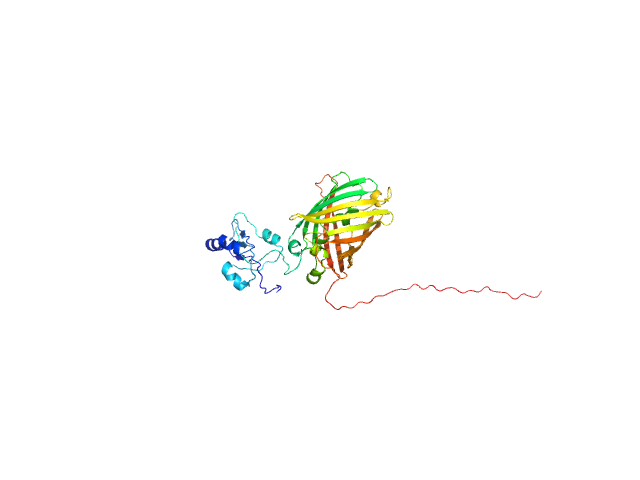
|
| Sample: |
DCV-GFP-St monomer, 41 kDa synthetic construct protein
|
| Buffer: |
20 mM Tris, 150 mM NaCL, 3% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2024 Mar 18
|
Programmable Protein-DNA Composite Nanostructures: from Nanostructure Construction to Protein-Induced Micro-Scale Material Self-Assembly and Functionalization.
Small 21(30):e2502060 (2025)
Zhou W, Strmšek Ž, Snoj J, Škarabot M, Jerala R
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.1 |
nm |
| VolumePorod |
54 |
nm3 |
|
|
|
|
|
|
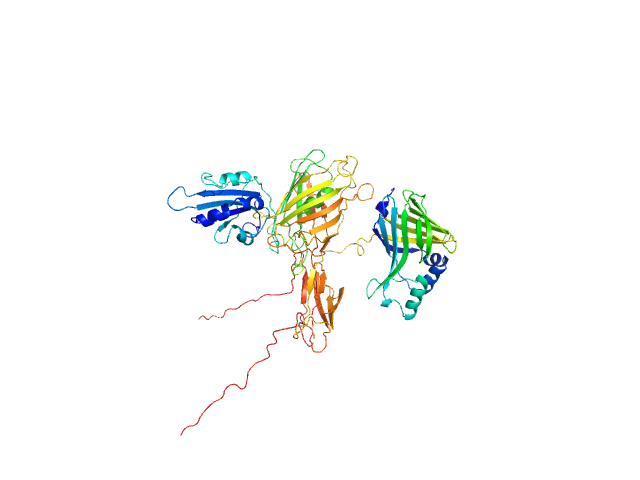
|
| Sample: |
DCV-GFP-St monomer, 41 kDa synthetic construct protein
NanoLuc-Sc monomer, 30 kDa synthetic construct protein
|
| Buffer: |
20 mM Tris, 150 mM NaCL, 3% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2024 Mar 18
|
Programmable Protein-DNA Composite Nanostructures: from Nanostructure Construction to Protein-Induced Micro-Scale Material Self-Assembly and Functionalization.
Small 21(30):e2502060 (2025)
Zhou W, Strmšek Ž, Snoj J, Škarabot M, Jerala R
|
| RgGuinier |
3.4 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
105 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Precursor microRNA 20a monomer, 23 kDa Homo sapiens RNA
|
| Buffer: |
50 mM potassium phosphate buffer, 1 mM MgCl2, 50 mM NaCl, 5 mM BME, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Oct 8
|
Structural features within precursor microRNA-20a regulate Dicer-TRBP processing
Journal of Molecular Biology :169317 (2025)
Liu Y, Harkner C, Westwood M, Munsayac A, Keane S
|
| RgGuinier |
3.1 |
nm |
| Dmax |
11.2 |
nm |
| VolumePorod |
32 |
nm3 |
|
|
|
|
|
|
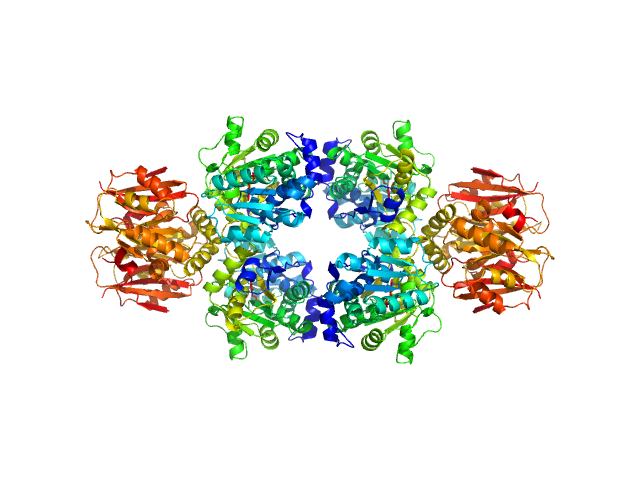
|
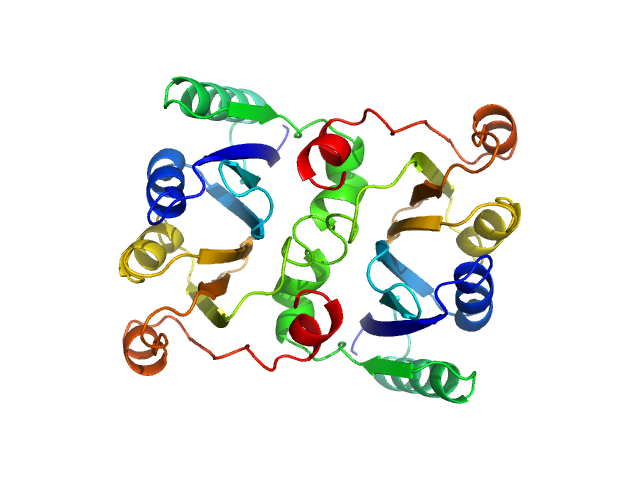
| Sample: |
L-threonine dehydratase biosynthetic IlvA tetramer, 233 kDa Escherichia coli (strain … protein
|
| Buffer: |
20 mM KPO4, 200 mM NaCl, 0.1 mM TCEP,, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Oct 14
|
Isoleucine Binding and Regulation of Escherichia coli and Staphylococcus aureus Threonine Dehydratase (IlvA).
Biochemistry (2025)
Yun MK, Subramanian C, Miller K, Jackson P, Radka CD, Rock CO
|
| RgGuinier |
4.5 |
nm |
| Dmax |
15.6 |
nm |
| VolumePorod |
284 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
L-threonine dehydratase biosynthetic IlvA Regulatory domain dimer, 46 kDa Escherichia coli (strain … protein
|
| Buffer: |
20 mM KPO4, 200 mM NaCl, 0.1 mM TCEP,, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Oct 14
|
Isoleucine Binding and Regulation of Escherichia coli and Staphylococcus aureus Threonine Dehydratase (IlvA).
Biochemistry (2025)
Yun MK, Subramanian C, Miller K, Jackson P, Radka CD, Rock CO
|
| RgGuinier |
2.3 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
50 |
nm3 |
|
|
|
|
|
|
|
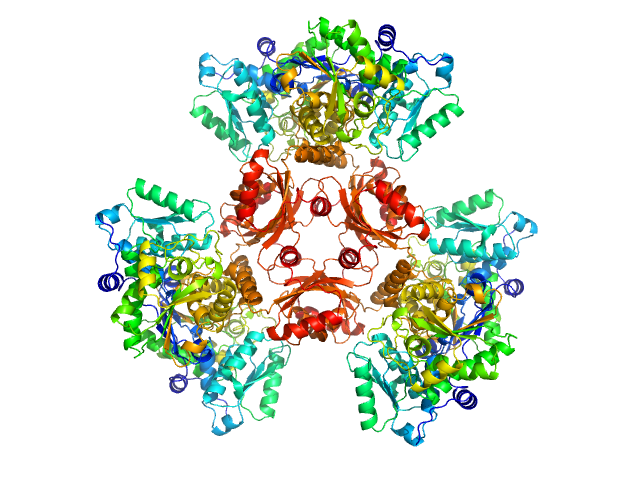
| Sample: |
L-threonine dehydratase biosynthetic IlvA hexamer, 288 kDa Staphylococcus aureus (strain … protein
|
| Buffer: |
20 mM KPO4, 200 mM NaCl, 0.1 mM TCEP,, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Oct 14
|
Isoleucine Binding and Regulation of Escherichia coli and Staphylococcus aureus Threonine Dehydratase (IlvA).
Biochemistry (2025)
Yun MK, Subramanian C, Miller K, Jackson P, Radka CD, Rock CO
|
| RgGuinier |
5.1 |
nm |
| Dmax |
17.1 |
nm |
| VolumePorod |
386 |
nm3 |
|
|
|
|
|
|
|
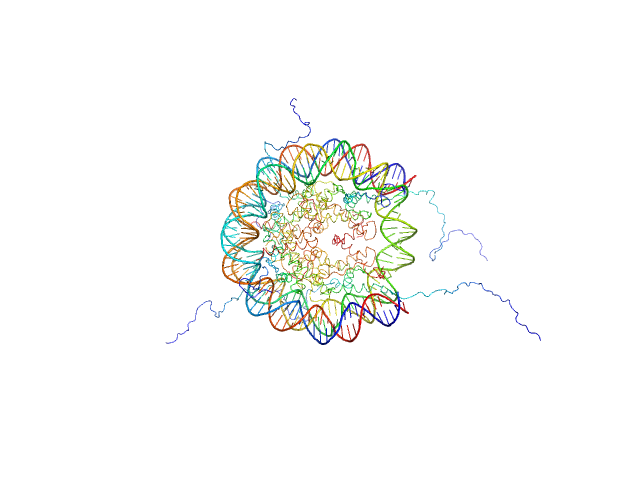
| Sample: |
Histone H3.2 dimer, 31 kDa Xenopus laevis protein
Histone H4 dimer, 23 kDa Xenopus laevis protein
Histone H2A dimer, 28 kDa Xenopus laevis protein
Histone H2B 1.1 dimer, 28 kDa Xenopus laevis protein
Widom 601 DNA sequence dimer, 89 kDa Escherichia coli DNA
|
| Buffer: |
20 mM Tris-HCl , 50 mM KCl, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2022 Dec 2
|
BCL7 proteins, novel subunits of the mammalian SWI/SNF complex, bind the nucleosome core particle
Asgar Abbas kazrani
|
| RgGuinier |
4.3 |
nm |
| Dmax |
12.3 |
nm |
| VolumePorod |
350 |
nm3 |
|
|
|
|
|
|
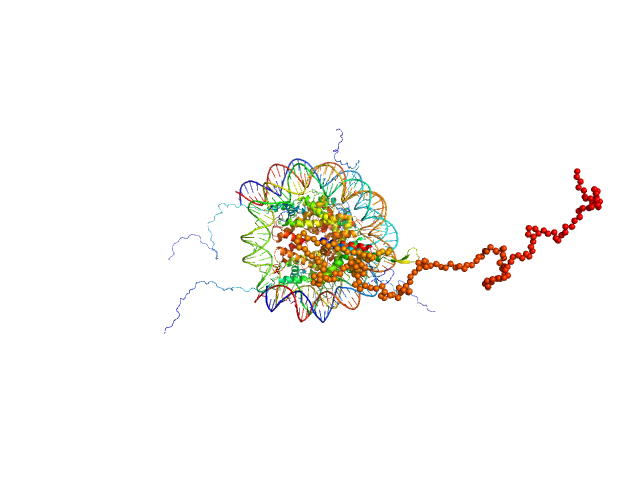
|
| Sample: |
Isoform 2 of B-cell CLL/lymphoma 7 protein family member A monomer, 25 kDa Homo sapiens protein
Histone H3.2 dimer, 31 kDa Xenopus laevis protein
Histone H4 dimer, 23 kDa Xenopus laevis protein
Histone H2A dimer, 28 kDa Xenopus laevis protein
Histone H2B 1.1 dimer, 28 kDa Xenopus laevis protein
Widom 601 DNA sequence dimer, 89 kDa Escherichia coli DNA
|
| Buffer: |
20 mM Tris-HCl , 50 mM KCl, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2022 Dec 2
|
BCL7 proteins, novel subunits of the mammalian SWI/SNF complex, bind the nucleosome core particle
Asgar Abbas kazrani
|
| RgGuinier |
4.7 |
nm |
| Dmax |
24.4 |
nm |
| VolumePorod |
400 |
nm3 |
|
|