|
|
|
|
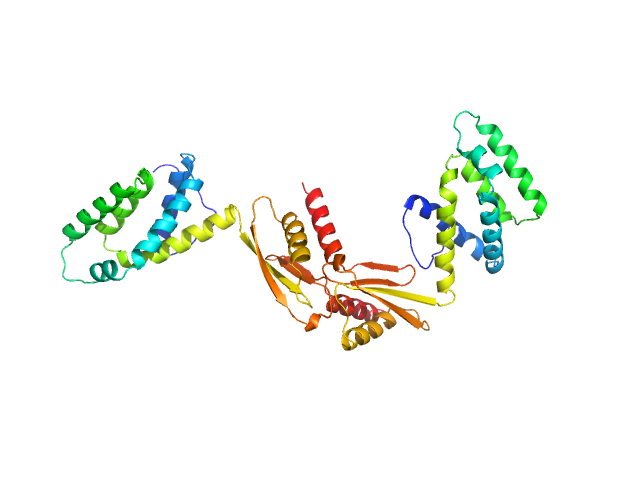
|
| Sample: |
ATP synthase subunit O, mitochondrial dimer, 42 kDa Homo sapiens protein
|
| Buffer: |
50 mM Na2HPO4, 300 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Mar 15
|
The OSCP Subunit of ATP Synthase is a Dimer in Solution: Strategy to Induce the Monomeric Protein as a New Tool for Drug Discovery.
J Mol Biol 437(17):169267 (2025)
Fabbian S, Gabbatore L, Morbiato L, Zotti M, Giorgio V, Battisutta R, Giachin G, Sosic A, Bellanda M
|
| RgGuinier |
3.7 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
78 |
nm3 |
|
|
|
|
|
|
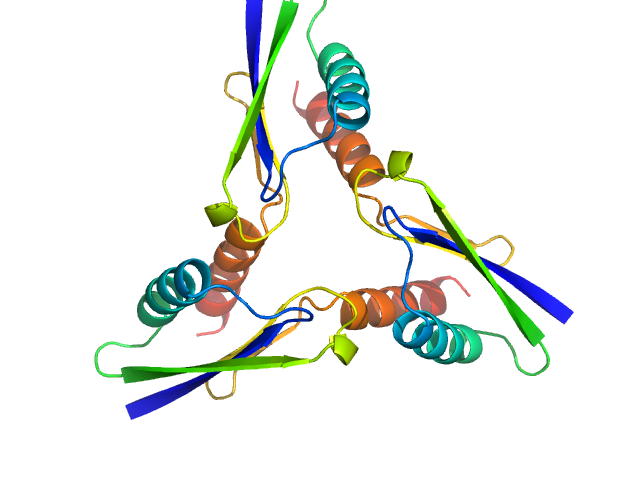
|
| Sample: |
ATP synthase subunit O, mitochondrial C-terminal section trimer, 25 kDa Homo sapiens protein
|
| Buffer: |
50 mM Na2HPO4, 300 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Mar 15
|
The OSCP Subunit of ATP Synthase is a Dimer in Solution: Strategy to Induce the Monomeric Protein as a New Tool for Drug Discovery.
J Mol Biol 437(17):169267 (2025)
Fabbian S, Gabbatore L, Morbiato L, Zotti M, Giorgio V, Battisutta R, Giachin G, Sosic A, Bellanda M
|
| RgGuinier |
2.1 |
nm |
| Dmax |
6.5 |
nm |
| VolumePorod |
32 |
nm3 |
|
|
|
|
|
|
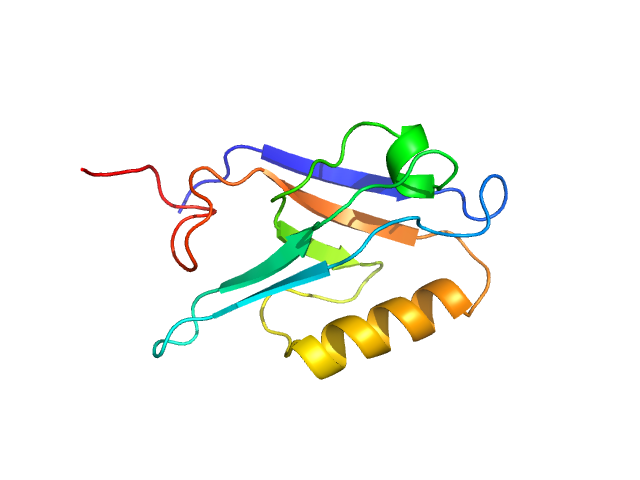
|
| Sample: |
Segment polarity protein dishevelled homolog DVL-3 monomer, 12 kDa Homo sapiens protein
|
| Buffer: |
50 mM sodium phosphate, 50 mM NaCl, 0.5 mM EDTA, pH: 6.5 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-2000, CEITEC on 2021 Jan 27
|
A class III ligand oscillates between internal and terminal binding modes as it engages with the Dishevelled PDZ domain.
Structure (2025)
Kumar J, Micka M, Komárek J, Klumpler T, Bystrý V, Sprangers R, Bařinka C, Bryja V, Tripsianes K
|
| RgGuinier |
1.6 |
nm |
| Dmax |
4.3 |
nm |
| VolumePorod |
20 |
nm3 |
|
|
|
|
|
|
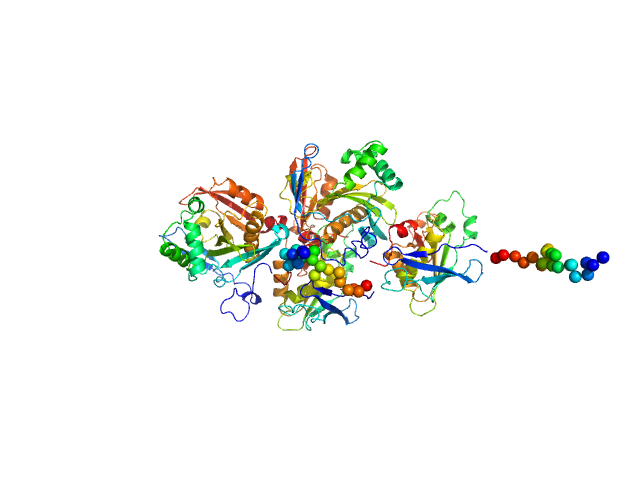
|
| Sample: |
N(5)-hydroxyornithine:cis-anhydromevalonyl coenzyme A-N(5)-transacylase sidF (Δ444-462) dimer, 107 kDa Aspergillus fumigatus (strain … protein
|
| Buffer: |
50 mM Tris, 200 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpoint 2.0, Institute of Biotechnology, Czech Academy of Sciences/Centre of Molecular Structure on 2023 Jan 18
|
SidF, a dual substrate N5-acetyl-N5-hydroxy-L-ornithine transacetylase involved in Aspergillus fumigatus siderophore biosynthesis
Journal of Structural Biology: X 11:100119 (2025)
Poonsiri T, Stransky J, Demitri N, Haas H, Cianci M, Benini S
|
| RgGuinier |
3.5 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
189 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Peptidase S9 tetramer, 304 kDa Mycolicibacterium phlei DSM … protein
|
| Buffer: |
10 mM Tris-HCl, 135 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BL-18, INDUS-2 on 2024 Mar 21
|
Structural and Functional Investigation of Putative Peptidase from Mycolicibacterium phlei
: An Exclusive Endopeptidase among S9C Subfamily
Biochemistry (2025)
Chandravanshi K, Jamdar S, Singh R, Kumar A, Mahato A, Agrawal R, Kumar S, Kumar A, Makde R
|
| RgGuinier |
5.2 |
nm |
| Dmax |
15.8 |
nm |
| VolumePorod |
559 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Peptidase S9 tetramer, 304 kDa Mycolicibacterium phlei DSM … protein
|
| Buffer: |
10 mM Tris-HCl, 135 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BL-18, INDUS-2 on 2024 Mar 21
|
Structural and Functional Investigation of Putative Peptidase from Mycolicibacterium phlei
: An Exclusive Endopeptidase among S9C Subfamily
Biochemistry (2025)
Chandravanshi K, Jamdar S, Singh R, Kumar A, Mahato A, Agrawal R, Kumar S, Kumar A, Makde R
|
| RgGuinier |
5.0 |
nm |
| Dmax |
15.7 |
nm |
| VolumePorod |
546 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Peptidase S9 tetramer, 304 kDa Mycolicibacterium phlei DSM … protein
|
| Buffer: |
10 mM Tris-HCl, 135 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BL-18, INDUS-2 on 2024 Mar 21
|
Structural and Functional Investigation of Putative Peptidase from Mycolicibacterium phlei
: An Exclusive Endopeptidase among S9C Subfamily
Biochemistry (2025)
Chandravanshi K, Jamdar S, Singh R, Kumar A, Mahato A, Agrawal R, Kumar S, Kumar A, Makde R
|
| RgGuinier |
5.1 |
nm |
| Dmax |
16.2 |
nm |
| VolumePorod |
563 |
nm3 |
|
|
|
|
|
|
|
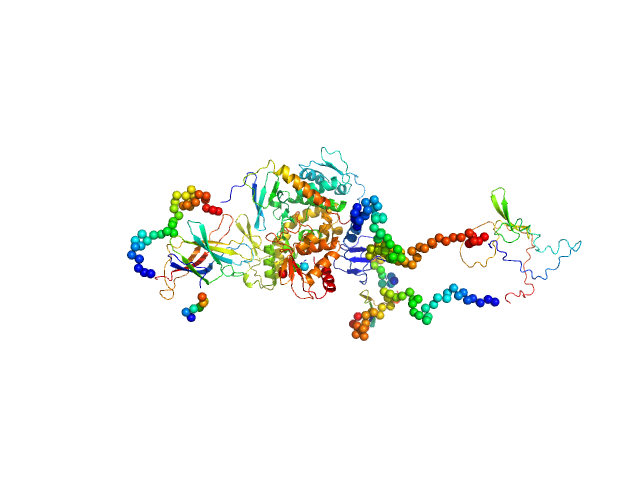
| Sample: |
Isoform 5 of E3 ubiquitin-protein ligase NEDD4-like monomer, 110 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1 mM TCEP, 1 mM EDTA, 3% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 Aug 17
|
Structural basis of ubiquitin ligase Nedd4-2 autoinhibition and regulation by calcium and 14-3-3 proteins
Nature Communications 16(1) (2025)
Janosev M, Kosek D, Tekel A, Joshi R, Honzejkova K, Pohl P, Obsil T, Obsilova V
|
| RgGuinier |
4.9 |
nm |
| Dmax |
22.0 |
nm |
| VolumePorod |
206 |
nm3 |
|
|
|
|
|
|
|
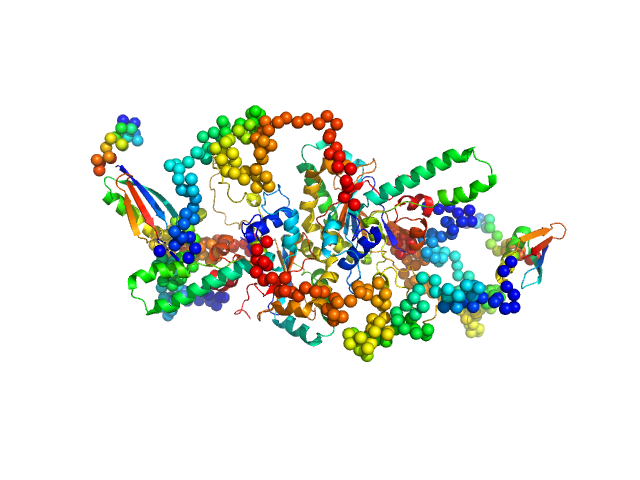
| Sample: |
Uncharacterized protein dimer, 122 kDa Ustilago maydis (strain … protein
|
| Buffer: |
20mM HEPES, 20 mM KCl, 200 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 Oct 25
|
Specific phosphoinositide interaction of Jps1 is a key feature during unconventional secretion in Ustilago maydis
Journal of Biological Chemistry :110215 (2025)
Dali S, Schultz M, Köster M, Kamel M, Busch M, Steinchen W, Hänsch S, Papadopoulos A, Reiners J, Smits S, Kedrov A, Altegoer F, Schipper K
|
| RgGuinier |
4.0 |
nm |
| Dmax |
14.7 |
nm |
| VolumePorod |
254 |
nm3 |
|
|
|
|
|
|
|
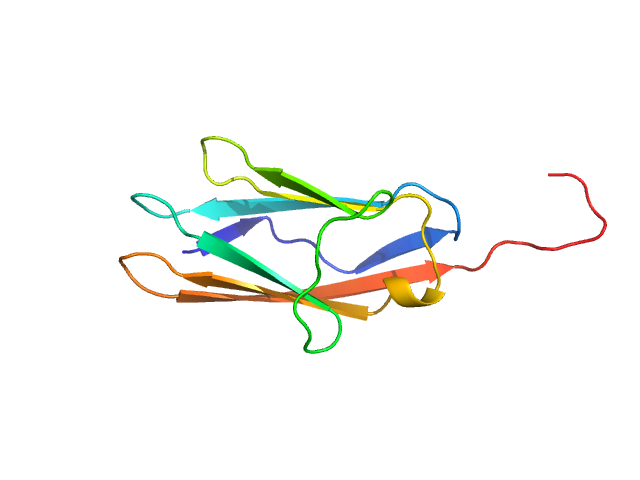
| Sample: |
Palladin monomer, 12 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES pH 7.4, 1 mM DTT, 100 mM NaCl, pH: |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2023 Aug 29
|
Integrated structural model of the palladin-actin complex using XL-MS, docking, NMR, and SAXS.
Protein Sci 34(5):e70122 (2025)
Sargent R, Liu DH, Yadav R, Glennenmeier D, Bradford C, Urbina N, Beck MR
|
| RgGuinier |
1.7 |
nm |
| Dmax |
6.8 |
nm |
| VolumePorod |
18 |
nm3 |
|
|