|
|
|
|
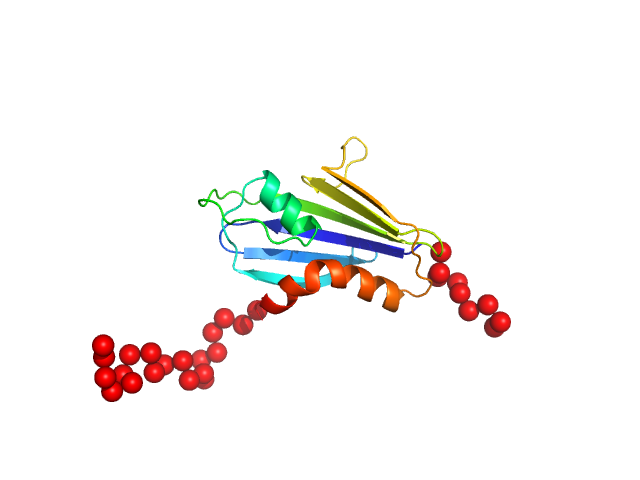
|
| Sample: |
Genome polyprotein monomer, 17 kDa Theiler's murine encephalomyelitis … protein
|
| Buffer: |
20 mM HEPES pH 7.4, 1 M KCl, 2% v/v glycerol, 1 mM DTT, pH: |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Apr 26
|
A new protein-dependent riboswitch activates ribosomal frameshifting
(2025)
Betts J, Jeffries C, Passchier T, Kung H, Graham S, Abdelhamid M, Howard J, Craggs T, Graham S, Brierley I, Leake M, Quinn S, Hill C
|
| RgGuinier |
1.9 |
nm |
| Dmax |
7.2 |
nm |
| VolumePorod |
30 |
nm3 |
|
|
|
|
|
|
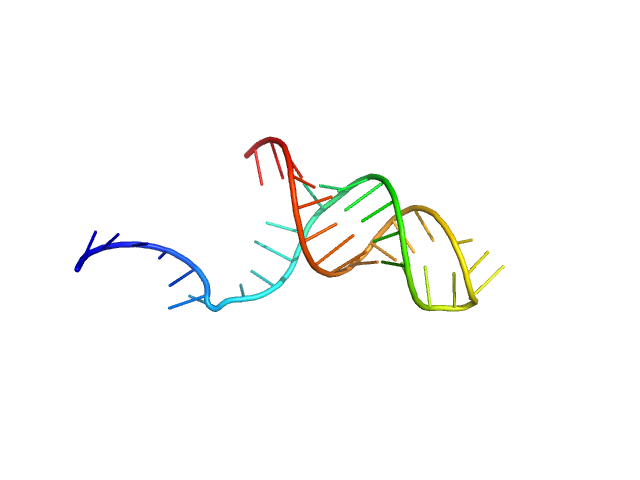
|
| Sample: |
Stimulatory element RNA monomer, 12 kDa synthetic RNA RNA
|
| Buffer: |
20 mM HEPES pH 7.4, 400 mM KCl 2% v/v glycerol, 1 mM DTT, pH: |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Apr 26
|
A new protein-dependent riboswitch activates ribosomal frameshifting
(2025)
Betts J, Jeffries C, Passchier T, Kung H, Graham S, Abdelhamid M, Howard J, Craggs T, Graham S, Brierley I, Leake M, Quinn S, Hill C
|
| RgGuinier |
1.9 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
19 |
nm3 |
|
|
|
|
|
|
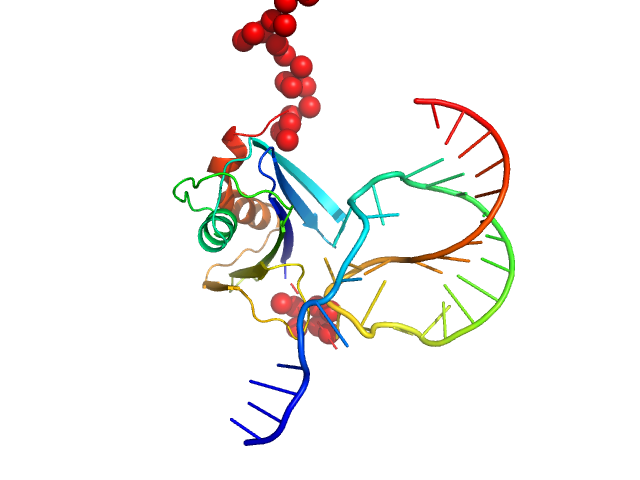
|
| Sample: |
Genome polyprotein monomer, 17 kDa Theiler's murine encephalomyelitis … protein
Stimulatory element RNA monomer, 12 kDa synthetic RNA RNA
|
| Buffer: |
20 mM HEPES pH 7.4, 400 mM KCl 2% v/v glycerol, 1 mM DTT, pH: |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Apr 26
|
A new protein-dependent riboswitch activates ribosomal frameshifting
(2025)
Betts J, Jeffries C, Passchier T, Kung H, Graham S, Abdelhamid M, Howard J, Craggs T, Graham S, Brierley I, Leake M, Quinn S, Hill C
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
46 |
nm3 |
|
|
|
|
|
|
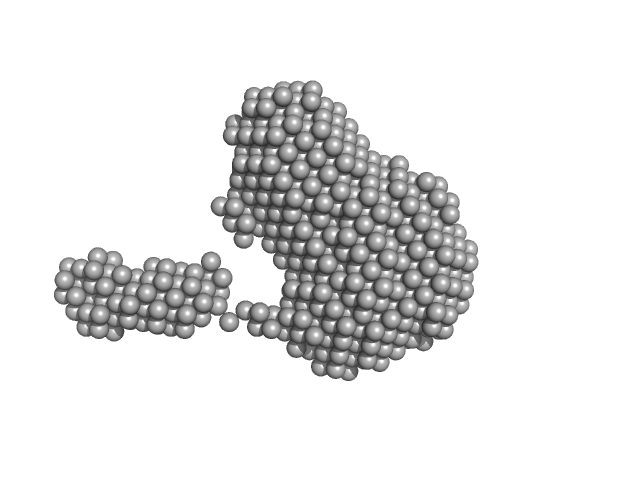
|
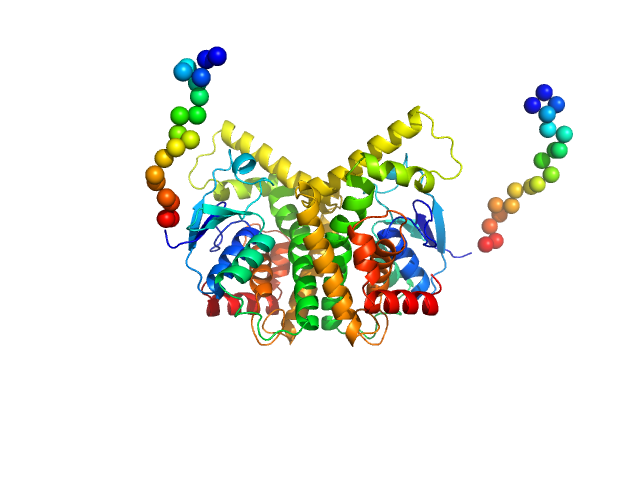
| Sample: |
Penicillin-Binding Protein 5 - PBP5∆1-39 monomer, 75 kDa Clostridioides difficile protein
|
| Buffer: |
100mM Tris·HCl, 150mM NaCl, 1mM EDTA, 1mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 May 13
|
SAXS of the Penicillin-Binding Protein 5 from Clostridioides difficile
Guillem Hernandez
|
| RgGuinier |
3.8 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
109 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
GST N-terminal domain-containing protein dimer, 60 kDa Gordonia rubripertincta protein
|
| Buffer: |
10 mM Tris, 300 mM NaCl,, pH: 8 |
| Experiment: |
SAXS
data collected at Xenocs Xeuss 2.0 Q-Xoom, Center for Structural Studies, Heinrich-Heine-University on 2022 Jan 20
|
Glutathione S
-Transferase Mediated Epoxide Conversion: Functional and Structural Properties of an Enantioselective Catalyst
ACS Catalysis :12308-12324 (2025)
Haarmann M, Scholz M, Lienkamp A, Burnik J, Justesen B, Reiners J, Maier A, Eggerichs D, Vocke K, Smits S, Günther Pomorski T, Hofmann E, Tischler D
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.2 |
nm |
| VolumePorod |
91 |
nm3 |
|
|
|
|
|
|
|
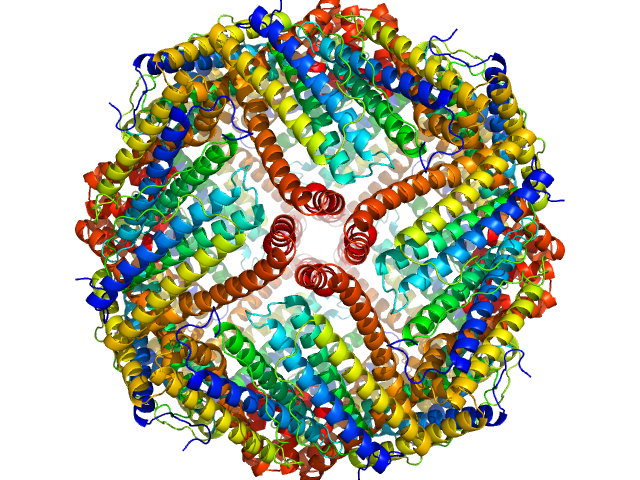
| Sample: |
Ferritin light chain 24-mer, 480 kDa Equus caballus protein
|
| Buffer: |
phosphate buffered saline, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 Jun 19
|
AF4-to-SAXS: expanded characterization of nanoparticles and proteins at the P12 BioSAXS beamline.
J Synchrotron Radiat (2025)
Da Vela S, Bartels K, Franke D, Soloviov D, Gräwert T, Molodenskiy D, Kolb B, Wilhelmy C, Drexel R, Meier F, Haas H, Langguth P, Graewert MA
|
| RgGuinier |
5.4 |
nm |
| Dmax |
13.3 |
nm |
| VolumePorod |
632 |
nm3 |
|
|
|
|
|
|
|
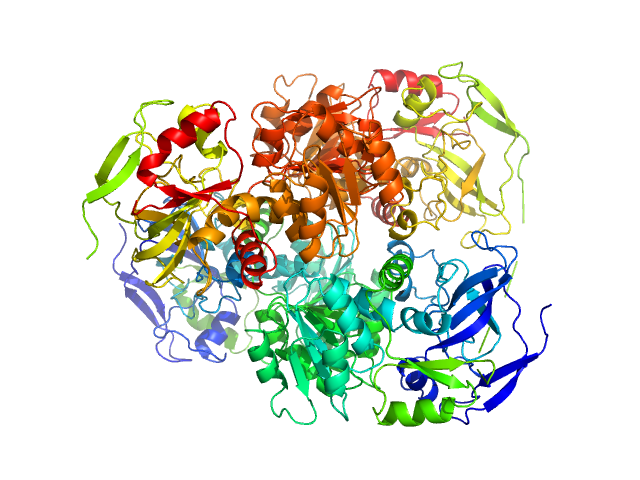
| Sample: |
Alcohol dehydrogenase 1 tetramer, 147 kDa Saccharomyces cerevisiae (strain … protein
|
| Buffer: |
phosphate buffered saline, 1% glycerol, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 Apr 13
|
AF4-to-SAXS: expanded characterization of nanoparticles and proteins at the P12 BioSAXS beamline.
J Synchrotron Radiat (2025)
Da Vela S, Bartels K, Franke D, Soloviov D, Gräwert T, Molodenskiy D, Kolb B, Wilhelmy C, Drexel R, Meier F, Haas H, Langguth P, Graewert MA
|
| RgGuinier |
3.4 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
197 |
nm3 |
|
|
|
|
|
|
|
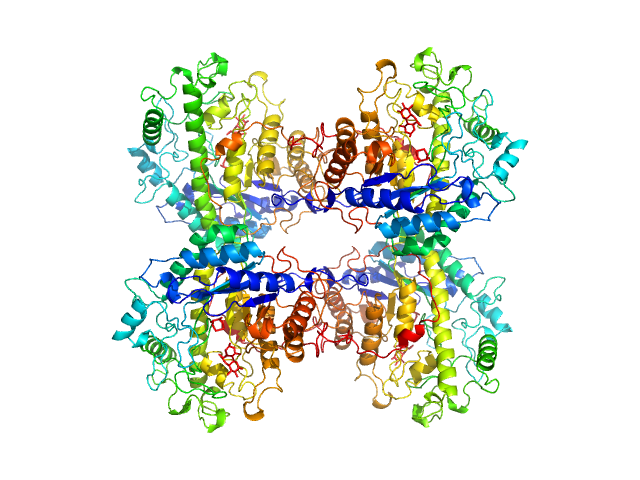
| Sample: |
Beta-amylase tetramer, 224 kDa Ipomoea batatas protein
|
| Buffer: |
phosphate buffered saline, 1% glycerol, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 Apr 13
|
AF4-to-SAXS: expanded characterization of nanoparticles and proteins at the P12 BioSAXS beamline.
J Synchrotron Radiat (2025)
Da Vela S, Bartels K, Franke D, Soloviov D, Gräwert T, Molodenskiy D, Kolb B, Wilhelmy C, Drexel R, Meier F, Haas H, Langguth P, Graewert MA
|
| RgGuinier |
4.2 |
nm |
| Dmax |
12.6 |
nm |
| VolumePorod |
297 |
nm3 |
|
|
|
|
|
|
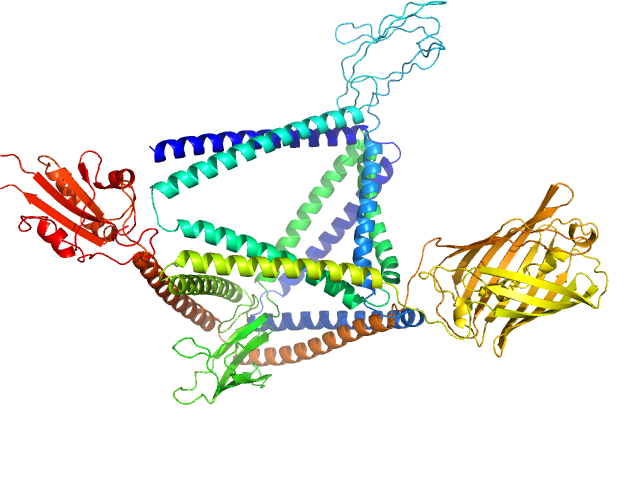
|
| Sample: |
TET12-2Sc-RFP-DCV monomer, 109 kDa synthetic construct protein
|
| Buffer: |
20 mM Tris, 150 mM NaCL, 3% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2024 Sep 24
|
Programmable Protein-DNA Composite Nanostructures: from Nanostructure Construction to Protein-Induced Micro-Scale Material Self-Assembly and Functionalization.
Small 21(30):e2502060 (2025)
Zhou W, Strmšek Ž, Snoj J, Škarabot M, Jerala R
|
| RgGuinier |
4.4 |
nm |
| Dmax |
13.6 |
nm |
| VolumePorod |
300 |
nm3 |
|
|
|
|
|
|
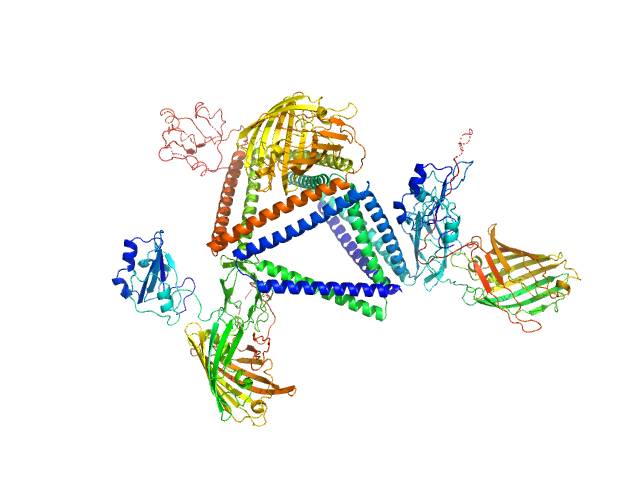
|
| Sample: |
TET12-2Sc-RFP-DCV monomer, 109 kDa synthetic construct protein
DCV-RFP-St monomer, 41 kDa synthetic construct protein
DCV-RFP-St monomer, 41 kDa synthetic construct protein
|
| Buffer: |
20 mM Tris, 150 mM NaCL, 3% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2024 Sep 24
|
Programmable Protein-DNA Composite Nanostructures: from Nanostructure Construction to Protein-Induced Micro-Scale Material Self-Assembly and Functionalization.
Small 21(30):e2502060 (2025)
Zhou W, Strmšek Ž, Snoj J, Škarabot M, Jerala R
|
| RgGuinier |
5.1 |
nm |
| Dmax |
15.4 |
nm |
| VolumePorod |
480 |
nm3 |
|
|