|
|
|
|
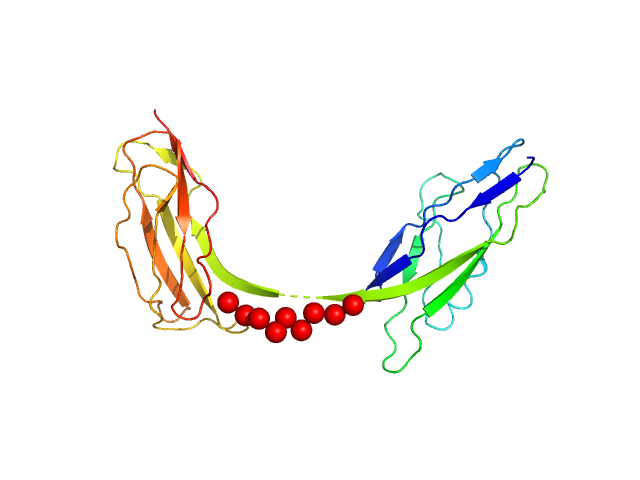
|
| Sample: |
Intimin, D00-D0 domain (6xHis tagged) monomer, 23 kDa Escherichia coli O127:H6 … protein
|
| Buffer: |
10 mM HEPES, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Jul 12
|
The extracellular juncture domains in the intimin passenger adopt a constitutively extended conformation inducing restraints to its sphere of action.
Sci Rep 10(1):21249 (2020)
Weikum J, Kulakova A, Tesei G, Yoshimoto S, Jægerum LV, Schütz M, Hori K, Skepö M, Harris P, Leo JC, Morth JP
|
| RgGuinier |
3.1 |
nm |
| Dmax |
8.3 |
nm |
| VolumePorod |
32 |
nm3 |
|
|
|
|
|
|
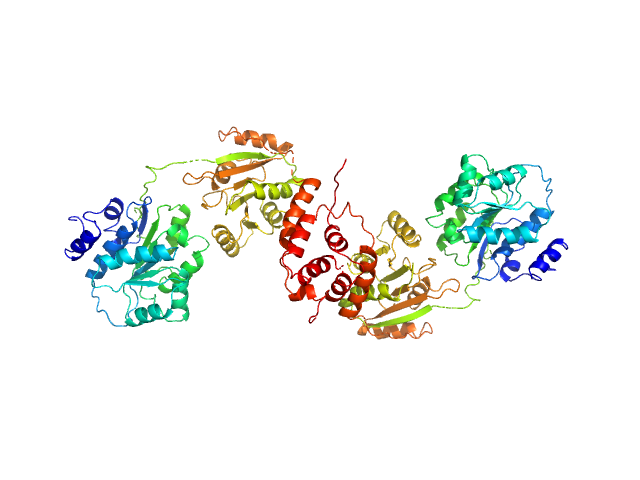
|
| Sample: |
Nucleolar RNA helicase 2 fragment 186-620 dimer, 98 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 500 mM NaCl, 10 % Glycerol, 2 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Mar 4
|
The Human RNA Helicase DDX21 Presents a Dimerization Interface Necessary for Helicase Activity
iScience 23(12):101811 (2020)
Marcaida M, Kauzlaric A, Duperrex A, Sülzle J, Moncrieffe M, Adebajo D, Manley S, Trono D, Dal Peraro M
|
| RgGuinier |
4.4 |
nm |
| Dmax |
16.9 |
nm |
| VolumePorod |
150 |
nm3 |
|
|
|
|
|
|
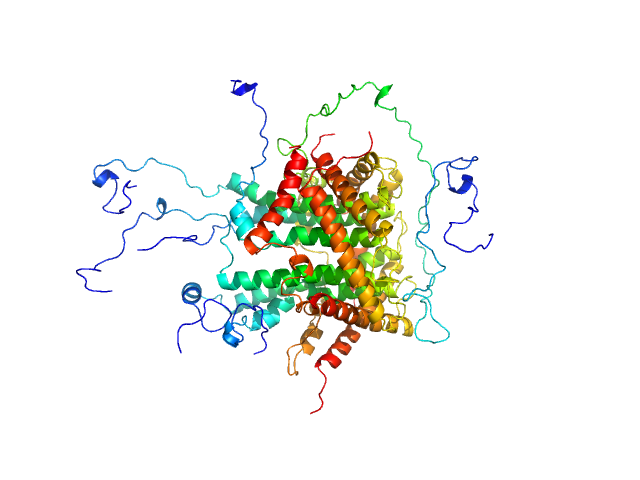
|
| Sample: |
Mitochondrial import inner membrane translocase subunit TIM8 trimer, 29 kDa Saccharomyces cerevisiae protein
Mitochondrial import inner membrane translocase subunit TIM13 trimer, 34 kDa Saccharomyces cerevisiae protein
Mitochondrial import inner membrane translocase subunit TIM23 monomer, 24 kDa Saccharomyces cerevisiae protein
|
| Buffer: |
50 mM Tris, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Apr 24
|
Structural basis of client specificity in mitochondrial membrane-protein chaperones.
Sci Adv 6(51) (2020)
Sučec I, Wang Y, Dakhlaoui O, Weinhäupl K, Jores T, Costa D, Hessel A, Brennich M, Rapaport D, Lindorff-Larsen K, Bersch B, Schanda P
|
| RgGuinier |
3.1 |
nm |
| Dmax |
11.3 |
nm |
|
|
|
|
|
|
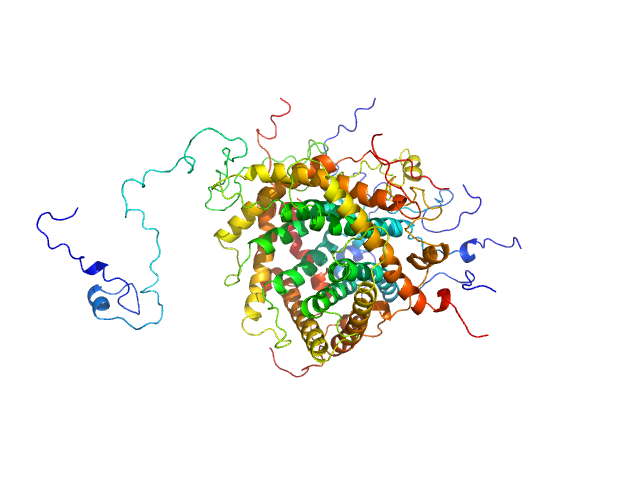
|
| Sample: |
Mitochondrial import inner membrane translocase subunit TIM9 trimer, 31 kDa Saccharomyces cerevisiae protein
Mitochondrial import inner membrane translocase subunit TIM10 trimer, 31 kDa Saccharomyces cerevisiae protein
Mitochondrial import inner membrane translocase subunit TIM23 monomer, 24 kDa Saccharomyces cerevisiae protein
|
| Buffer: |
50 mM Tris, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 May 18
|
Structural basis of client specificity in mitochondrial membrane-protein chaperones.
Sci Adv 6(51) (2020)
Sučec I, Wang Y, Dakhlaoui O, Weinhäupl K, Jores T, Costa D, Hessel A, Brennich M, Rapaport D, Lindorff-Larsen K, Bersch B, Schanda P
|
| RgGuinier |
3.2 |
nm |
| Dmax |
7.7 |
nm |
| VolumePorod |
146 |
nm3 |
|
|
|
|
|
|
|
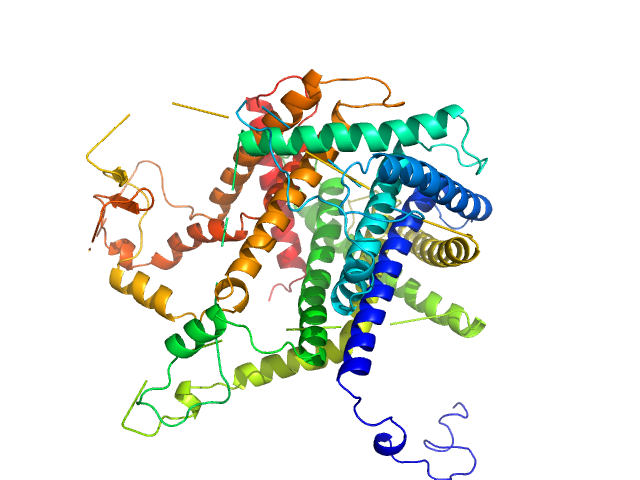
| Sample: |
Mitochondrial import inner membrane translocase subunit TIM8 trimer, 29 kDa Saccharomyces cerevisiae protein
Mitochondrial import inner membrane translocase subunit TIM13 trimer, 34 kDa Saccharomyces cerevisiae protein
|
| Buffer: |
50 mM Tris, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Apr 21
|
Structural basis of client specificity in mitochondrial membrane-protein chaperones.
Sci Adv 6(51) (2020)
Sučec I, Wang Y, Dakhlaoui O, Weinhäupl K, Jores T, Costa D, Hessel A, Brennich M, Rapaport D, Lindorff-Larsen K, Bersch B, Schanda P
|
| RgGuinier |
3.1 |
nm |
| Dmax |
10.9 |
nm |
| VolumePorod |
136 |
nm3 |
|
|
|
|
|
|
|
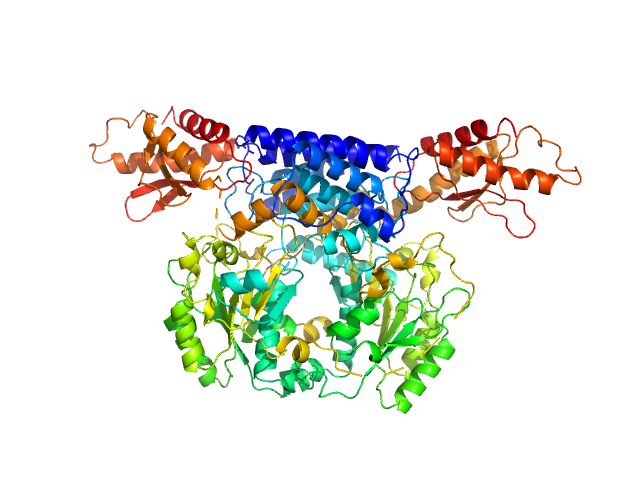
| Sample: |
Cysteine sulfinic acid decarboxylase dimer, 117 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 200 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2018 Feb 4
|
Structure and substrate specificity determinants of the taurine biosynthetic enzyme cysteine sulphinic acid decarboxylase.
J Struct Biol :107674 (2020)
Mahootchi E, Raasakka A, Luan W, Muruganandam G, Loris R, Haavik J, Kursula P
|
| RgGuinier |
3.4 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
198 |
nm3 |
|
|
|
|
|
|
|
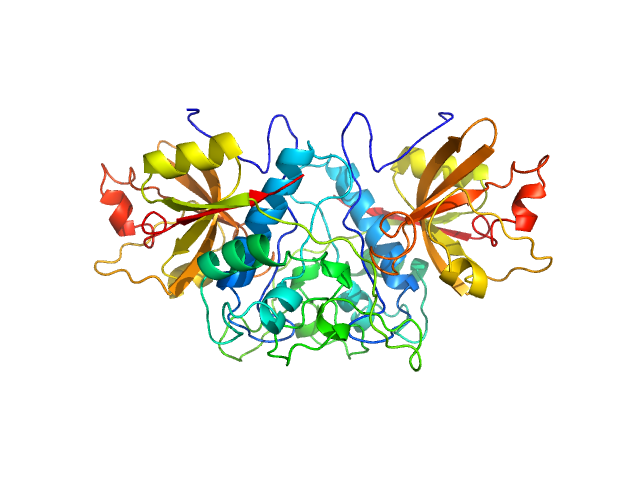
| Sample: |
Cathepsin Z dimer, 54 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium acetate, 1 mM EDTA, pH: 5.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Dec 6
|
Human cathepsin X/Z is a biologically active homodimer.
Biochim Biophys Acta Proteins Proteom :140567 (2020)
Dolenc I, Štefe I, Turk D, Taler-Verčič A, Turk B, Turk V, Stoka V
|
| RgGuinier |
2.6 |
nm |
| Dmax |
6.5 |
nm |
| VolumePorod |
89 |
nm3 |
|
|
|
|
|
|
|
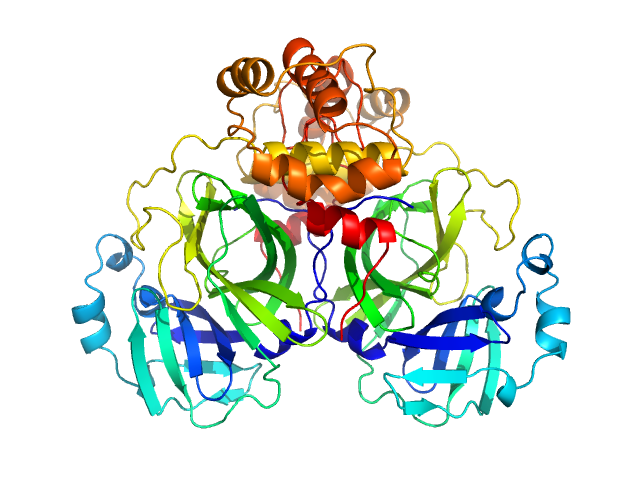
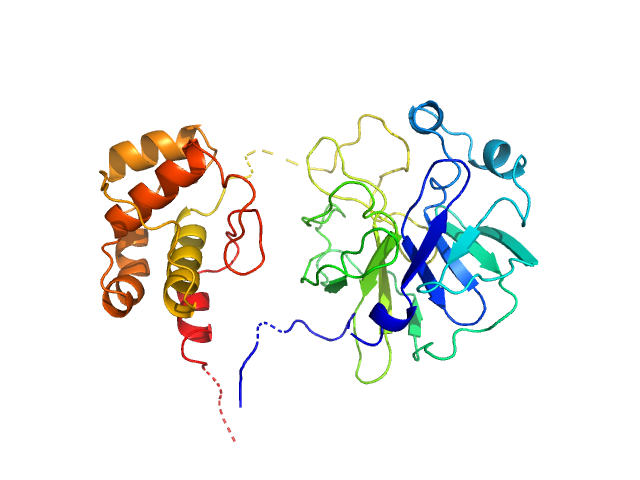
| Sample: |
3C-like proteinase from SARS-CoV-2 replicase polyprotein 1a dimer, 68 kDa Severe acute respiratory … protein
|
| Buffer: |
50 mM Tris, 1 mM DTT, 1 mM EDTA, pH: 7.4 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-2000, University of British Columbia on 2020 Jun 1
|
Crystallographic structure of wild-type SARS-CoV-2 main protease acyl-enzyme intermediate with physiological C-terminal autoprocessing site.
Nat Commun 11(1):5877 (2020)
Lee J, Worrall LJ, Vuckovic M, Rosell FI, Gentile F, Ton AT, Caveney NA, Ban F, Cherkasov A, Paetzel M, Strynadka NCJ
|
| RgGuinier |
2.7 |
nm |
| Dmax |
8.8 |
nm |
| VolumePorod |
93 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
3C-like proteinase from SARS-CoV-2 replicase polyprotein 1a, PT9 mutant monomer, 34 kDa Severe acute respiratory … protein
|
| Buffer: |
50 mM Tris, 1 mM DTT, 1 mM EDTA, pH: 7.4 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-2000, University of British Columbia on 2020 Jun 1
|
Crystallographic structure of wild-type SARS-CoV-2 main protease acyl-enzyme intermediate with physiological C-terminal autoprocessing site.
Nat Commun 11(1):5877 (2020)
Lee J, Worrall LJ, Vuckovic M, Rosell FI, Gentile F, Ton AT, Caveney NA, Ban F, Cherkasov A, Paetzel M, Strynadka NCJ
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.2 |
nm |
| VolumePorod |
54 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Synthetic nanobody Sybody 23 monomer, 16 kDa synthetic construct protein
|
| Buffer: |
50 mM Tris 100 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 May 5
|
Selection, biophysical and structural analysis of synthetic nanobodies that effectively neutralize SARS-CoV-2
Nature Communications 11(1) (2020)
Custódio T, Das H, Sheward D, Hanke L, Pazicky S, Pieprzyk J, Sorgenfrei M, Schroer M, Gruzinov A, Jeffries C, Graewert M, Svergun D, Dobrev N, Remans K, Seeger M, McInerney G, Murrell B, Hällberg B, Löw C
|
| RgGuinier |
2.1 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
22 |
nm3 |
|
|