|
|
|
|

|
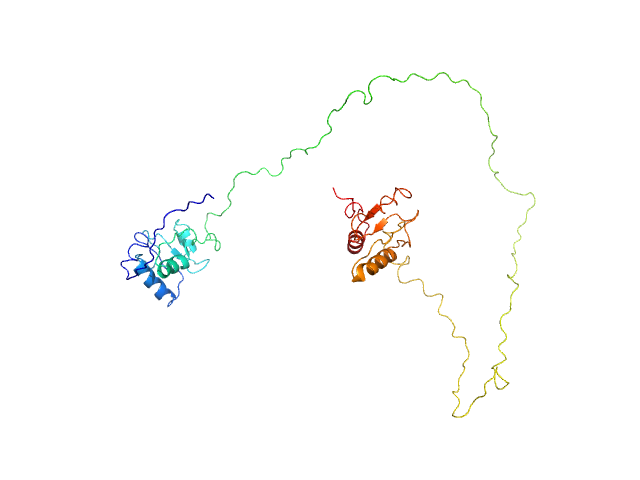
| Sample: |
ESX-1 secretion-associated protein EspK monomer, 30 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
20 mM Tris-HCl pH, 300 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Apr 12
|
Structural Analysis of the Partially Disordered Protein EspK from Mycobacterium Tuberculosis
Crystals 11(1):18 (2020)
Gijsbers A, Sánchez-Puig N, Gao Y, Peters P, Ravelli R, Siliqi D
|
| RgGuinier |
2.2 |
nm |
| Dmax |
8.4 |
nm |
| VolumePorod |
52 |
nm3 |
|
|
|
|
|
|
|
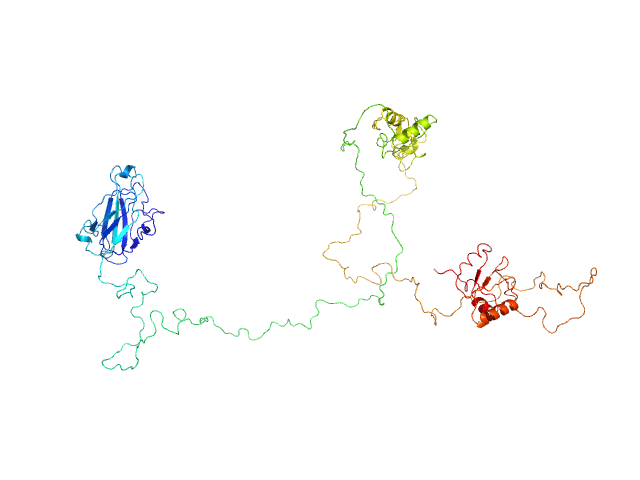
| Sample: |
DNA repair protein XRCC1ΔN dimer, 76 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris-HCl pH 7.5, 150 mM NaCl, 10% glycerol, 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2011 Jul 15
|
An atypical BRCT-BRCT interaction with the XRCC1 scaffold protein compacts human DNA Ligase IIIα within a flexible DNA repair complex.
Nucleic Acids Res (2020)
Hammel M, Rashid I, Sverzhinsky A, Pourfarjam Y, Tsai MS, Ellenberger T, Pascal JM, Kim IK, Tainer JA, Tomkinson AE
|
| RgGuinier |
5.4 |
nm |
| Dmax |
19.5 |
nm |
| VolumePorod |
170 |
nm3 |
|
|
|
|
|
|
|
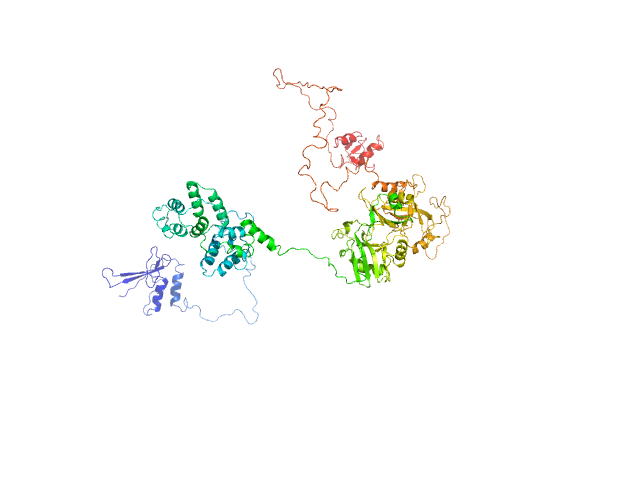
| Sample: |
DNA repair protein XRCC1, 69 kDa Homo sapiens protein
|
| Buffer: |
200 mM NaCl, 20 mM Tris-HCl, pH 7.5, 2% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Jun 19
|
An atypical BRCT-BRCT interaction with the XRCC1 scaffold protein compacts human DNA Ligase IIIα within a flexible DNA repair complex.
Nucleic Acids Res (2020)
Hammel M, Rashid I, Sverzhinsky A, Pourfarjam Y, Tsai MS, Ellenberger T, Pascal JM, Kim IK, Tainer JA, Tomkinson AE
|
| RgGuinier |
6.3 |
nm |
| Dmax |
26.9 |
nm |
| VolumePorod |
330 |
nm3 |
|
|
|
|
|
|
|
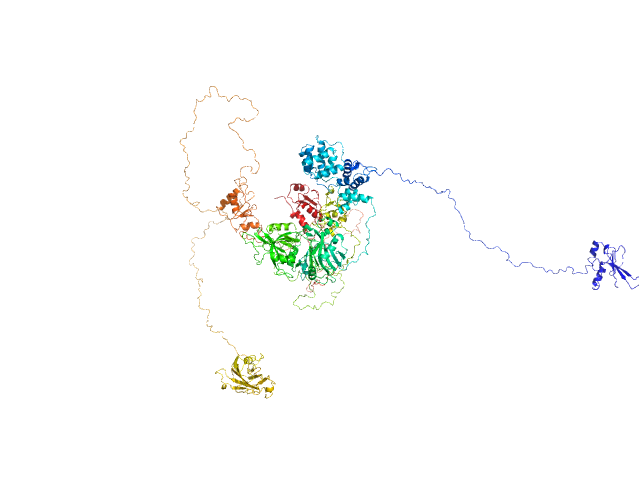
| Sample: |
DNA ligase 3 (DNA ligase III alpha), Homo sapiens protein
|
| Buffer: |
25 mM Tris-HCl pH 7.5, 150 mM NaCl, 10% glycerol, 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2011 Mar 31
|
An atypical BRCT-BRCT interaction with the XRCC1 scaffold protein compacts human DNA Ligase IIIα within a flexible DNA repair complex.
Nucleic Acids Res (2020)
Hammel M, Rashid I, Sverzhinsky A, Pourfarjam Y, Tsai MS, Ellenberger T, Pascal JM, Kim IK, Tainer JA, Tomkinson AE
|
| RgGuinier |
6.1 |
nm |
| Dmax |
21.0 |
nm |
| VolumePorod |
240 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
DNA repair protein XRCC1, 69 kDa Homo sapiens protein
DNA ligase 3 (DNA ligase III alpha), Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 5 mM MgCl₂, 0.2 mM PMSF, 1 mM benzamidine, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Aug 18
|
An atypical BRCT-BRCT interaction with the XRCC1 scaffold protein compacts human DNA Ligase IIIα within a flexible DNA repair complex.
Nucleic Acids Res (2020)
Hammel M, Rashid I, Sverzhinsky A, Pourfarjam Y, Tsai MS, Ellenberger T, Pascal JM, Kim IK, Tainer JA, Tomkinson AE
|
| RgGuinier |
6.2 |
nm |
| Dmax |
27.9 |
nm |
| VolumePorod |
675 |
nm3 |
|
|
|
|
|
|
|
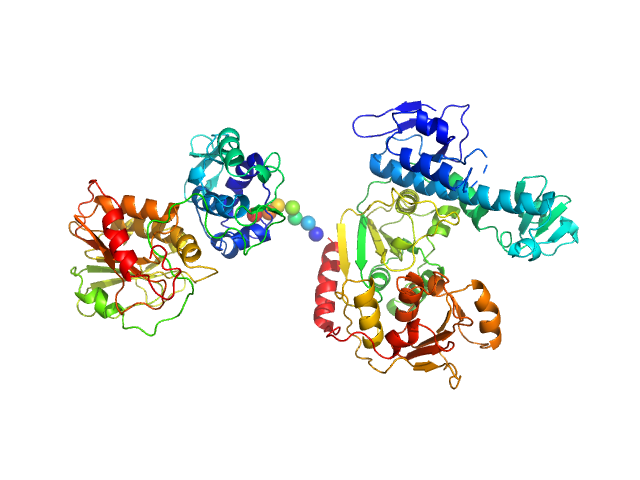
| Sample: |
NsP2 protein monomer, 90 kDa Chikungunya virus protein
|
| Buffer: |
20 mM Hepes pH 7.4, 150 mM NaCl, 1 mM DTT, 5% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at 23A, Taiwan Photon Source, NSRRC on 2019 Aug 10
|
Inter-domain Flexibility of Chikungunya Virus nsP2 Helicase-Protease Differentially Influences Viral RNA Replication and Infectivity.
J Virol (2020)
Law YS, Wang S, Tan YB, Shih O, Utt A, Goh WY, Lian BJ, Chen MW, Jeng US, Merits A, Luo D
|
| RgGuinier |
3.9 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
134 |
nm3 |
|
|
|
|
|
|
|
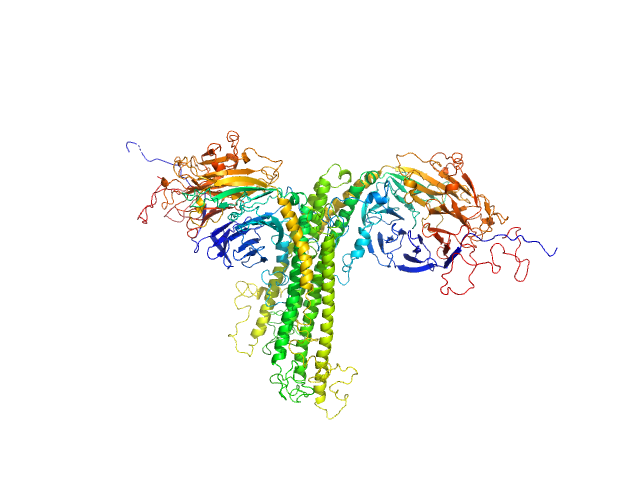
| Sample: |
Anaphase Promoting Complex/Cyclosome Subunit 4 dimer, 184 kDa Arabidopsis thaliana protein
|
| Buffer: |
10 mM Tris 150 mM NaCl 1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Mar 6
|
Structural Characterisation of the Arabidopsis thaliana APC4
Steven De Gieter
|
| RgGuinier |
4.6 |
nm |
| Dmax |
16.2 |
nm |
| VolumePorod |
259 |
nm3 |
|
|
|
|
|
|
|
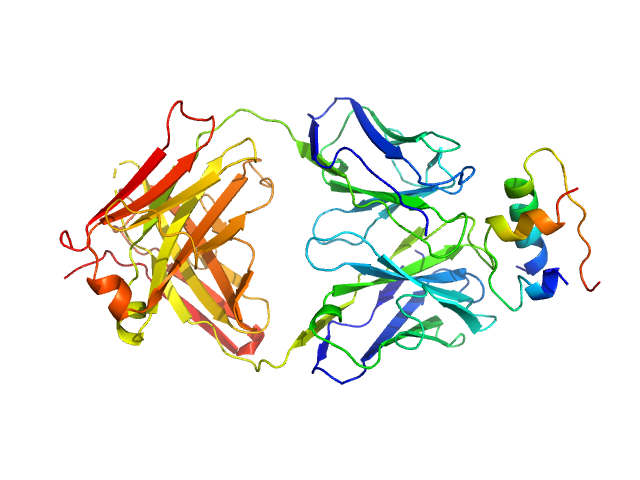
| Sample: |
Insulin (Insulin B chain and Insulin A chain) monomer, 6 kDa Homo sapiens protein
HUI-018 Fab monomer, 47 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 140 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at I911-4, MAX IV on 2013 Mar 7
|
Insulin Binding to the Analytical Antibody Sandwich Pair OXI
‐005 and HUI
‐018 – Epitope Mapping and Binding Properties
Protein Science (2020)
Johansson E, Wu X, Yu B, Yang Z, Cao Z, Wiberg C, Jeppesen C, Poulsen F
|
| RgGuinier |
3.4 |
nm |
| Dmax |
10.7 |
nm |
| VolumePorod |
86 |
nm3 |
|
|
|
|
|
|
|
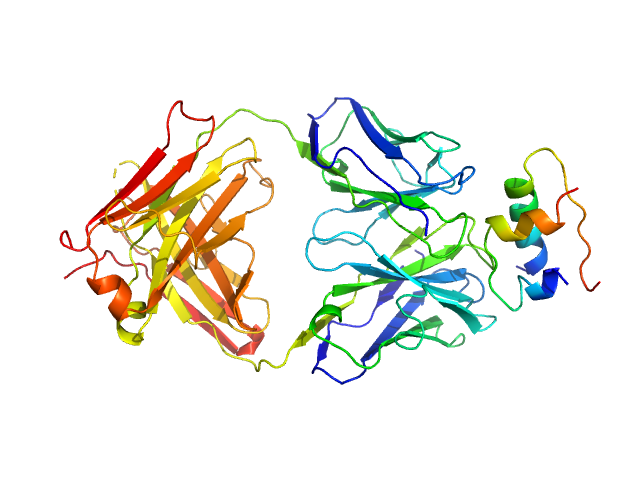
| Sample: |
Insulin (Insulin B chain and Insulin A chain) monomer, 6 kDa Homo sapiens protein
HUI-018 Fab monomer, 47 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 140 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at I911-4, MAX IV on 2013 Mar 7
|
Insulin Binding to the Analytical Antibody Sandwich Pair OXI
‐005 and HUI
‐018 – Epitope Mapping and Binding Properties
Protein Science (2020)
Johansson E, Wu X, Yu B, Yang Z, Cao Z, Wiberg C, Jeppesen C, Poulsen F
|
| RgGuinier |
3.7 |
nm |
| Dmax |
13.4 |
nm |
| VolumePorod |
105 |
nm3 |
|
|
|
|
|
|
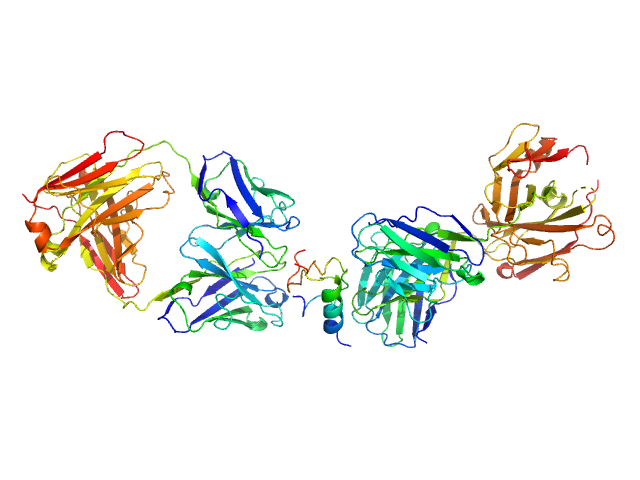
|
| Sample: |
HUI-018 Fab monomer, 47 kDa Mus musculus protein
Insulin (Insulin B chain and Insulin A chain) monomer, 6 kDa Sus scrofa protein
OXI-005 Fab monomer, 47 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 140 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-2000, Novo Nordisk A/S on 2019 Mar 28
|
Insulin Binding to the Analytical Antibody Sandwich Pair OXI
‐005 and HUI
‐018 – Epitope Mapping and Binding Properties
Protein Science (2020)
Johansson E, Wu X, Yu B, Yang Z, Cao Z, Wiberg C, Jeppesen C, Poulsen F
|
| RgGuinier |
4.5 |
nm |
| Dmax |
14.2 |
nm |
| VolumePorod |
138 |
nm3 |
|
|