|
|
|
|
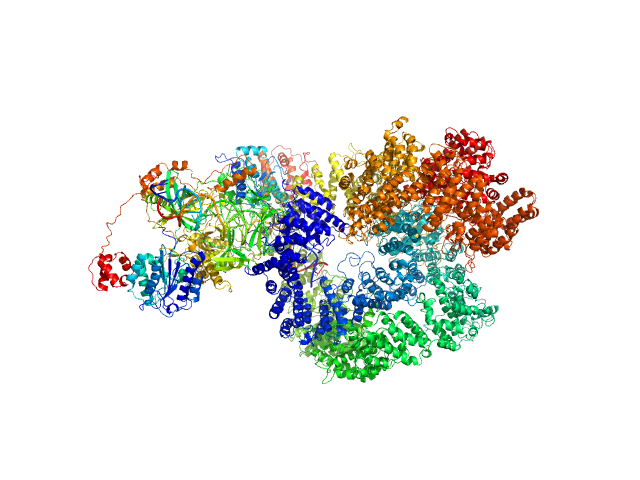
|
| Sample: |
X-ray repair cross-complementing protein 6 monomer, 70 kDa Homo sapiens protein
X-ray repair cross-complementing protein 5 monomer, 83 kDa Homo sapiens protein
DNA-dependent protein kinase catalytic subunit monomer, 468 kDa Homo sapiens protein
DsDNA dimer, 21 kDa DNA
|
| Buffer: |
50 mM Tris-HCl, 100 mM NaCl, 5% glycerol, 0.01% sodium azide, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Dec 30
|
Visualizing functional dynamicity in the DNA-dependent protein kinase holoenzyme DNA-PK complex by integrating SAXS with cryo-EM.
Prog Biophys Mol Biol (2020)
Hammel M, Rosenberg DJ, Bierma J, Hura GL, Lees-Miller SP, Tainer JA
|
| RgGuinier |
6.5 |
nm |
| Dmax |
23.1 |
nm |
| VolumePorod |
1090 |
nm3 |
|
|
|
|
|
|
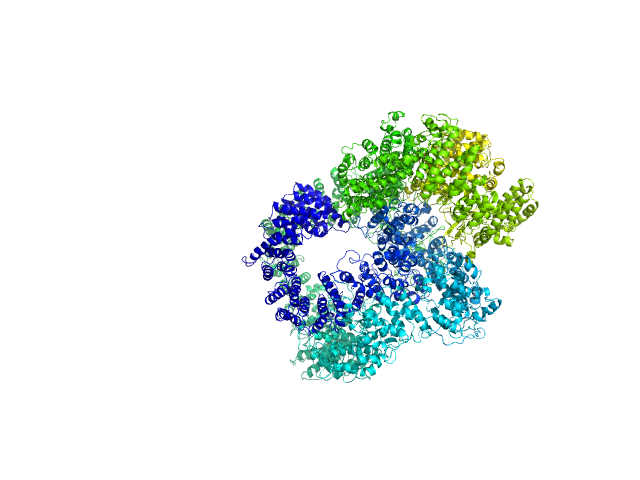
|
| Sample: |
X-ray repair cross-complementing protein 6 monomer, 70 kDa Homo sapiens protein
X-ray repair cross-complementing protein 5 monomer, 83 kDa Homo sapiens protein
DNA-dependent protein kinase catalytic subunit monomer, 468 kDa Homo sapiens protein
DsDNA dimer, 21 kDa DNA
|
| Buffer: |
50 mM Tris-HCl, 100 mM NaCl, 5% glycerol, 0.01% sodium azide, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Dec 30
|
Visualizing functional dynamicity in the DNA-dependent protein kinase holoenzyme DNA-PK complex by integrating SAXS with cryo-EM.
Prog Biophys Mol Biol (2020)
Hammel M, Rosenberg DJ, Bierma J, Hura GL, Lees-Miller SP, Tainer JA
|
| RgGuinier |
7.5 |
nm |
| Dmax |
29.4 |
nm |
| VolumePorod |
1440 |
nm3 |
|
|
|
|
|
|
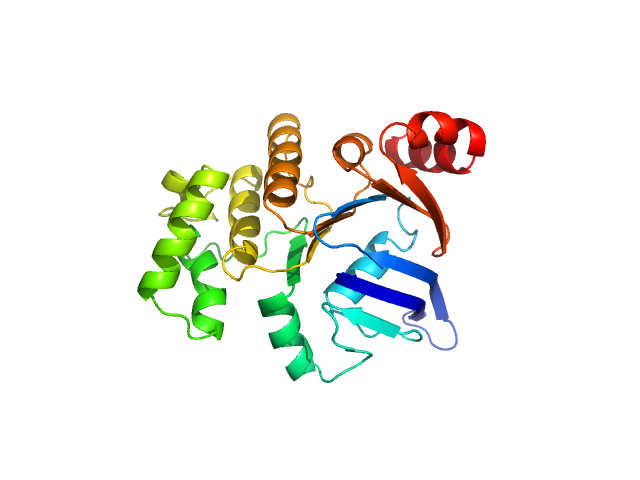
|
| Sample: |
ABC transporter, ATP-binding protein (Nucleotide-Binding Domain SaNsrF) monomer, 31 kDa Streptococcus agalactiae protein
|
| Buffer: |
100 mM HEPES, 150 mM NaCl, 10% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at Xenocs Xeuss 2.0 Q-Xoom, Center for Structural Studies, Heinrich-Heine-University on 2019 Dec 11
|
Characterization of the nucleotide-binding domain NsrF from the BceAB-type ABC-transporter NsrFP from the human pathogen Streptococcus agalactiae
Scientific Reports 10(1) (2020)
Furtmann F, Porta N, Hoang D, Reiners J, Schumacher J, Gottstein J, Gohlke H, Smits S
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.9 |
nm |
| VolumePorod |
64 |
nm3 |
|
|
|
|
|
|
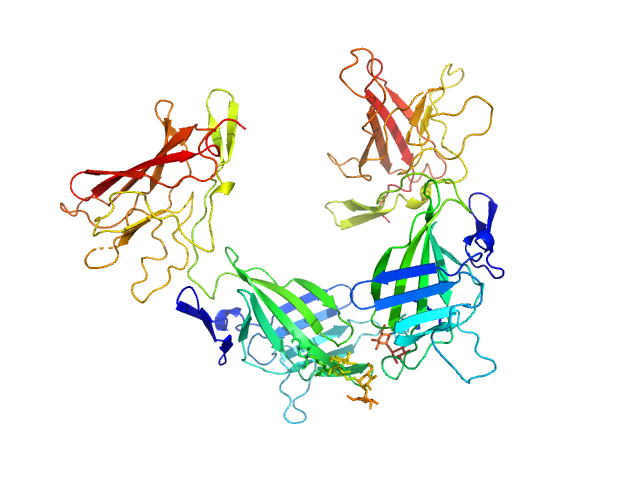
|
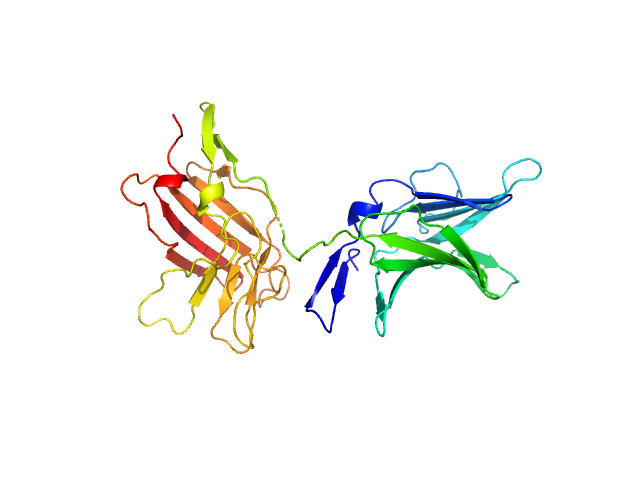
| Sample: |
Cation-independent mannose-6-phosphate receptor dimer, 67 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2020 Jan 25
|
Structure of the Human Cation-Independent Mannose 6-Phosphate/IGF2 Receptor Domains 7–11 Uncovers the Mannose 6-Phosphate Binding Site of Domain 9
Structure (2020)
Bochel A, Williams C, McCoy A, Hoppe H, Winter A, Nicholls R, Harlos K, Jones E, Berger I, Hassan A, Crump M
|
| RgGuinier |
3.3 |
nm |
| Dmax |
9.1 |
nm |
| VolumePorod |
88 |
nm3 |
|
|
|
|
|
|
|
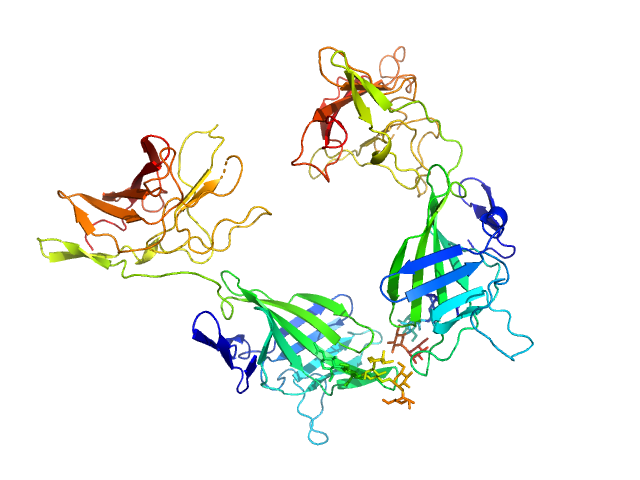
| Sample: |
Cation-independent mannose-6-phosphate receptor monomer, 34 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2020 Mar 3
|
Structure of the Human Cation-Independent Mannose 6-Phosphate/IGF2 Receptor Domains 7–11 Uncovers the Mannose 6-Phosphate Binding Site of Domain 9
Structure (2020)
Bochel A, Williams C, McCoy A, Hoppe H, Winter A, Nicholls R, Harlos K, Jones E, Berger I, Hassan A, Crump M
|
| RgGuinier |
2.5 |
nm |
| Dmax |
7.8 |
nm |
| VolumePorod |
48 |
nm3 |
|
|
|
|
|
|
|
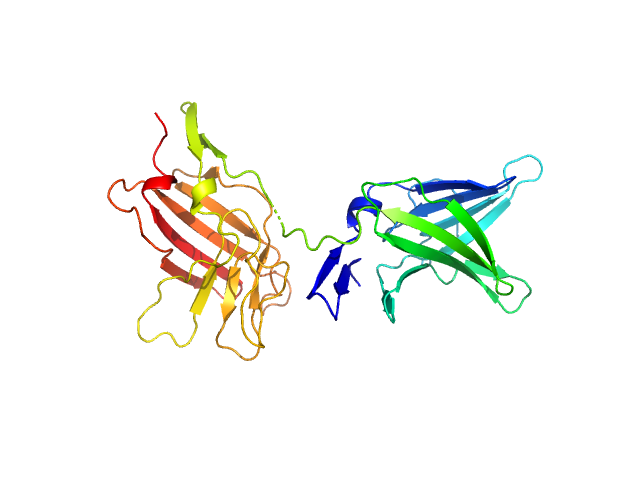
| Sample: |
Cation-independent mannose-6-phosphate receptor dimer, 67 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2020 Jan 25
|
Structure of the Human Cation-Independent Mannose 6-Phosphate/IGF2 Receptor Domains 7–11 Uncovers the Mannose 6-Phosphate Binding Site of Domain 9
Structure (2020)
Bochel A, Williams C, McCoy A, Hoppe H, Winter A, Nicholls R, Harlos K, Jones E, Berger I, Hassan A, Crump M
|
| RgGuinier |
3.3 |
nm |
| Dmax |
9.2 |
nm |
| VolumePorod |
93 |
nm3 |
|
|
|
|
|
|
|
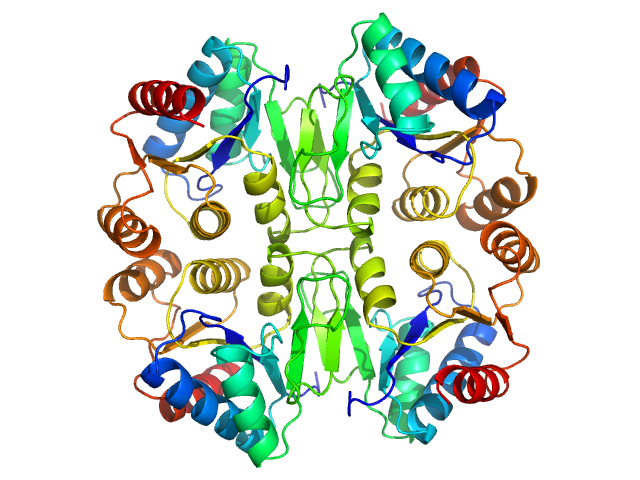
| Sample: |
Cation-independent mannose-6-phosphate receptor monomer, 34 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2020 Mar 3
|
Structure of the Human Cation-Independent Mannose 6-Phosphate/IGF2 Receptor Domains 7–11 Uncovers the Mannose 6-Phosphate Binding Site of Domain 9
Structure (2020)
Bochel A, Williams C, McCoy A, Hoppe H, Winter A, Nicholls R, Harlos K, Jones E, Berger I, Hassan A, Crump M
|
| RgGuinier |
2.5 |
nm |
| Dmax |
7.8 |
nm |
| VolumePorod |
69 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
P450 cytochrome, putative (Moco carrier protein) tetramer, 75 kDa Rippkaea orientalis (strain … protein
|
| Buffer: |
100 mM Tris-HCl, 300 mM NaCl, 2 %(v/v) glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Feb 5
|
The structure of the Moco carrier protein from Rippkaea orientalis.
Acta Crystallogr F Struct Biol Commun 76(Pt 9):453-463 (2020)
Krausze J, Hercher TW, Archna A, Kruse T
|
| RgGuinier |
2.8 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
107 |
nm3 |
|
|
|
|
|
|
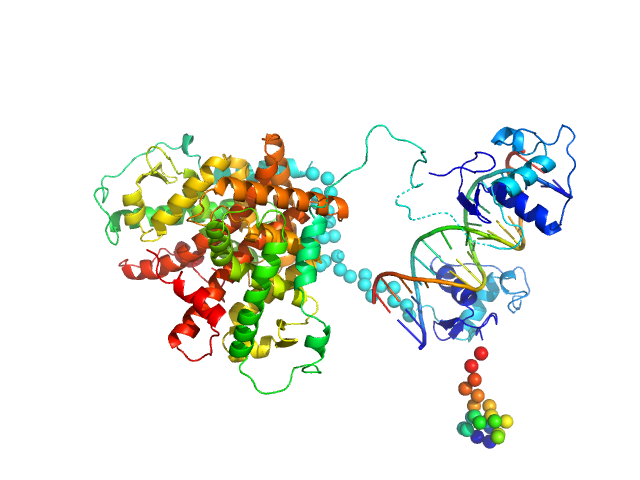
|
| Sample: |
Retinoic acid receptor alpha, RAR monomer, 41 kDa Mus musculus protein
Retinoic acid receptor RXR-alpha monomer, 38 kDa Mus musculus protein
DNA response element HoxB13 DR0 monomer, 10 kDa DNA
|
| Buffer: |
20 mM Tris, pH 8, 150 mM NaCl, 5% v/v glycerol, 1 mM CHAPS, 4 mM MgSO4, 1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2014 Jan 19
|
Structural basis for DNA recognition and allosteric control of the retinoic acid receptors RAR–RXR
Nucleic Acids Research (2020)
Osz J, McEwen A, Bourguet M, Przybilla F, Peluso-Iltis C, Poussin-Courmontagne P, Mély Y, Cianférani S, Jeffries C, Svergun D, Rochel N
|
| RgGuinier |
3.8 |
nm |
| Dmax |
14.5 |
nm |
| VolumePorod |
132 |
nm3 |
|
|
|
|
|
|
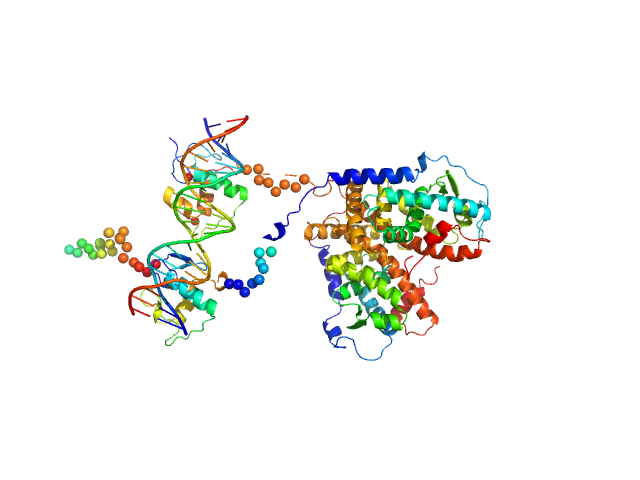
|
| Sample: |
Retinoic acid receptor alpha, RAR monomer, 41 kDa Mus musculus protein
Retinoic acid receptor RXR-alpha monomer, 38 kDa Mus musculus protein
DNA response element F11r DR5 monomer, 13 kDa DNA
|
| Buffer: |
20 mM Tris, pH 8, 150 mM NaCl, 5% v/v glycerol, 1 mM CHAPS, 4 mM MgSO4, 1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2014 Jan 19
|
Structural basis for DNA recognition and allosteric control of the retinoic acid receptors RAR–RXR
Nucleic Acids Research (2020)
Osz J, McEwen A, Bourguet M, Przybilla F, Peluso-Iltis C, Poussin-Courmontagne P, Mély Y, Cianférani S, Jeffries C, Svergun D, Rochel N
|
| RgGuinier |
4.0 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
130 |
nm3 |
|
|