|
|
|
|
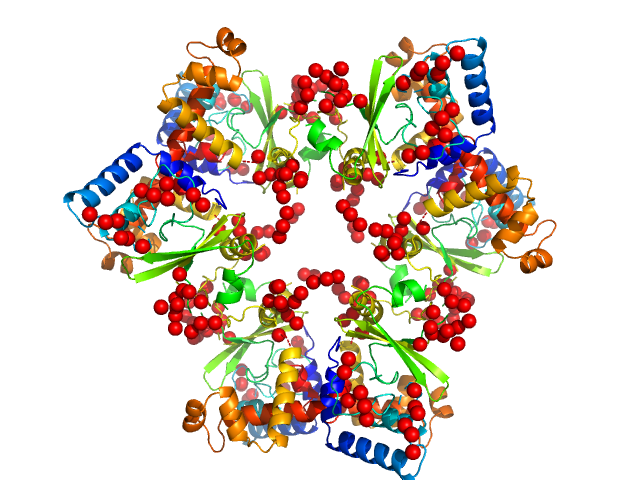
|
| Sample: |
Regulator Of Virulence interconnected with the Csr system hexamer, 175 kDa Yersinia pseudotuberculosis protein
|
| Buffer: |
50 mM TRIS pH= 8, 500 mM NaCl, 5 mM DTT, 5 % v/v glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Jun 18
|
RovC - a novel type of hexameric transcriptional activator promoting type VI secretion gene expression
PLOS Pathogens 16(9):e1008552 (2020)
Knittel V, Sadana P, Seekircher S, Stolle A, Körner B, Volk M, Jeffries C, Svergun D, Heroven A, Scrima A, Dersch P, Mecsas J
|
| RgGuinier |
4.2 |
nm |
| Dmax |
12.4 |
nm |
| VolumePorod |
280 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
C-terminal domain-like carotenoid protein dimer, 33 kDa Tolypothrix sp. PCC … protein
|
| Buffer: |
10 mM potassium phosphate, 100 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 May 15
|
Structural analysis of a new carotenoid-binding protein: the C-terminal domain homolog of the OCP
Scientific Reports 10(1) (2020)
Dominguez-Martin M, Hammel M, Gupta S, Lechno-Yossef S, Sutter M, Rosenberg D, Chen Y, Petzold C, Ralston C, Polívka T, Kerfeld C
|
| RgGuinier |
2.5 |
nm |
| Dmax |
9.1 |
nm |
| VolumePorod |
55 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
C-terminal domain-like carotenoid protein dimer, 36 kDa Tolypothrix sp. PCC … protein
|
| Buffer: |
10 mM potassium phosphate, 100 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 May 15
|
Structural analysis of a new carotenoid-binding protein: the C-terminal domain homolog of the OCP
Scientific Reports 10(1) (2020)
Dominguez-Martin M, Hammel M, Gupta S, Lechno-Yossef S, Sutter M, Rosenberg D, Chen Y, Petzold C, Ralston C, Polívka T, Kerfeld C
|
| RgGuinier |
2.5 |
nm |
| Dmax |
7.9 |
nm |
| VolumePorod |
62 |
nm3 |
|
|
|
|
|
|
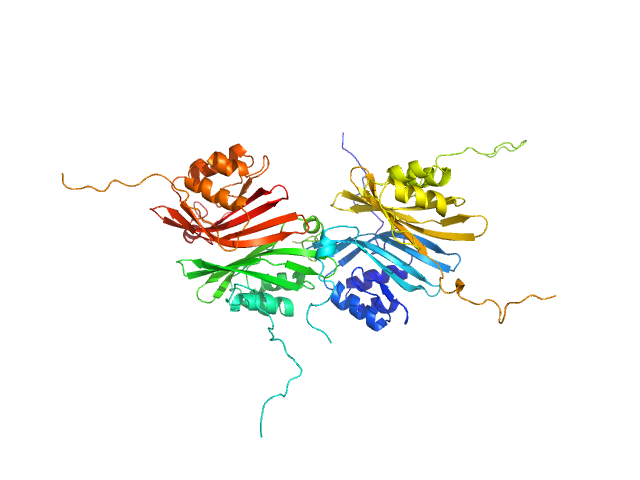
|
| Sample: |
C-terminal domain-like carotenoid protein tetramer, 66 kDa Tolypothrix sp. PCC … protein
|
| Buffer: |
10 mM potassium phosphate, 100 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 May 15
|
Structural analysis of a new carotenoid-binding protein: the C-terminal domain homolog of the OCP
Scientific Reports 10(1) (2020)
Dominguez-Martin M, Hammel M, Gupta S, Lechno-Yossef S, Sutter M, Rosenberg D, Chen Y, Petzold C, Ralston C, Polívka T, Kerfeld C
|
| RgGuinier |
3.2 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
110 |
nm3 |
|
|
|
|
|
|
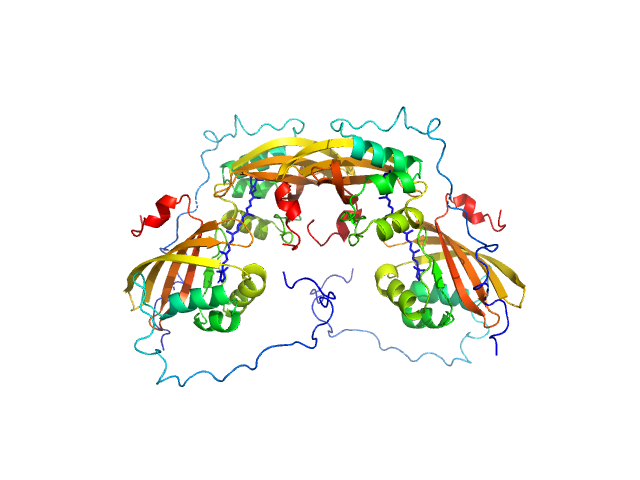
|
| Sample: |
C-terminal domain-like carotenoid protein tetramer, 73 kDa Tolypothrix sp. PCC … protein
|
| Buffer: |
10 mM potassium phosphate, 100 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 May 15
|
Structural analysis of a new carotenoid-binding protein: the C-terminal domain homolog of the OCP
Scientific Reports 10(1) (2020)
Dominguez-Martin M, Hammel M, Gupta S, Lechno-Yossef S, Sutter M, Rosenberg D, Chen Y, Petzold C, Ralston C, Polívka T, Kerfeld C
|
| RgGuinier |
3.3 |
nm |
| Dmax |
12.1 |
nm |
| VolumePorod |
157 |
nm3 |
|
|
|
|
|
|
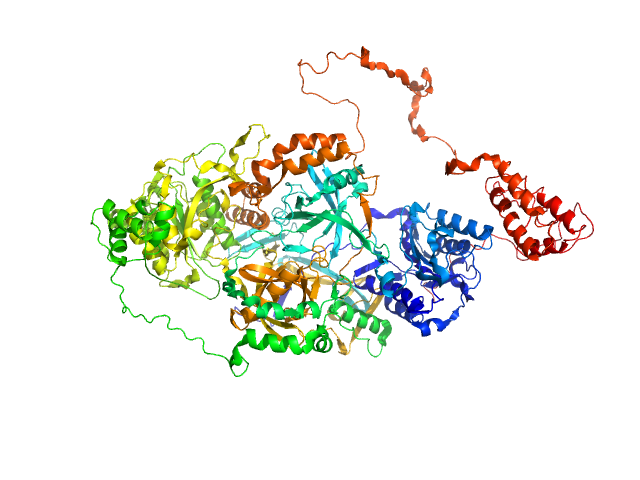
|
| Sample: |
X-ray repair cross-complementing protein 6 monomer, 70 kDa Homo sapiens protein
X-ray repair cross-complementing protein 5 monomer, 83 kDa Homo sapiens protein
|
| Buffer: |
50 mM Hepes, 50 mM KCl, 5 mM MgCl2, 5% glycerol and 0.2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Mar 12
|
Visualizing functional dynamicity in the DNA-dependent protein kinase holoenzyme DNA-PK complex by integrating SAXS with cryo-EM.
Prog Biophys Mol Biol (2020)
Hammel M, Rosenberg DJ, Bierma J, Hura GL, Lees-Miller SP, Tainer JA
|
| RgGuinier |
4.2 |
nm |
| Dmax |
14.9 |
nm |
| VolumePorod |
253 |
nm3 |
|
|
|
|
|
|
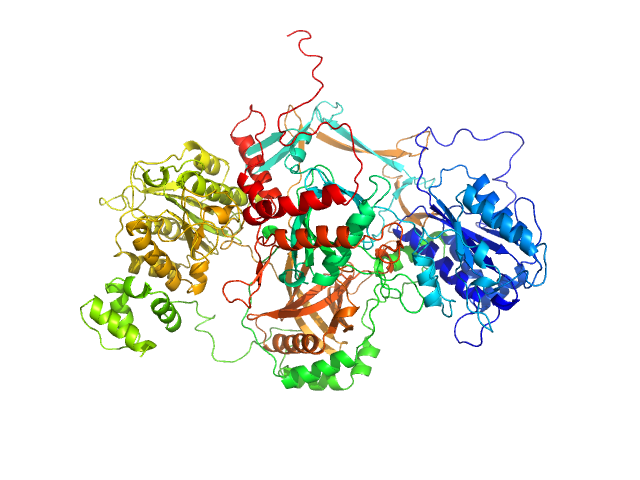
|
| Sample: |
X-ray repair cross-complementing protein 6 monomer, 70 kDa Homo sapiens protein
X-ray repair cross-complementing protein 5 ΔCTR monomer, 64 kDa Homo sapiens protein
|
| Buffer: |
50 mM Hepes, 50 mM KCl, 5 mM MgCl2, 5% glycerol and 0.2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Mar 12
|
Visualizing functional dynamicity in the DNA-dependent protein kinase holoenzyme DNA-PK complex by integrating SAXS with cryo-EM.
Prog Biophys Mol Biol (2020)
Hammel M, Rosenberg DJ, Bierma J, Hura GL, Lees-Miller SP, Tainer JA
|
| RgGuinier |
3.8 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
240 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
X-ray repair cross-complementing protein 6 monomer, 70 kDa Homo sapiens protein
X-ray repair cross-complementing protein 5 monomer, 83 kDa Homo sapiens protein
Y-DNA monomer, 18 kDa DNA
|
| Buffer: |
50 mM Tris-HCl, 100 mM NaCl, 5% glycerol, 0.01% sodium azide, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2010 Jan 8
|
Visualizing functional dynamicity in the DNA-dependent protein kinase holoenzyme DNA-PK complex by integrating SAXS with cryo-EM.
Prog Biophys Mol Biol (2020)
Hammel M, Rosenberg DJ, Bierma J, Hura GL, Lees-Miller SP, Tainer JA
|
| RgGuinier |
4.1 |
nm |
| Dmax |
14.8 |
nm |
| VolumePorod |
280 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
DNA-dependent protein kinase catalytic subunit monomer, 468 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCl, 100 mM NaCl, 5% glycerol, 0.01% sodium azide, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Dec 30
|
Visualizing functional dynamicity in the DNA-dependent protein kinase holoenzyme DNA-PK complex by integrating SAXS with cryo-EM.
Prog Biophys Mol Biol (2020)
Hammel M, Rosenberg DJ, Bierma J, Hura GL, Lees-Miller SP, Tainer JA
|
| RgGuinier |
5.6 |
nm |
| Dmax |
15.6 |
nm |
| VolumePorod |
1040 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
DNA-dependent protein kinase catalytic subunit monomer, 468 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCl, 100 mM KCl, 5% (v/v) glycerol, 0.2 mM EDTA containing 0.1 mM benzamidine, 0.2 mM PMSF and 0.2 µg/ml pepstatin, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2010 Jan 8
|
Visualizing functional dynamicity in the DNA-dependent protein kinase holoenzyme DNA-PK complex by integrating SAXS with cryo-EM.
Prog Biophys Mol Biol (2020)
Hammel M, Rosenberg DJ, Bierma J, Hura GL, Lees-Miller SP, Tainer JA
|
| RgGuinier |
5.5 |
nm |
| Dmax |
16.1 |
nm |
| VolumePorod |
918 |
nm3 |
|
|