|
|
|
|
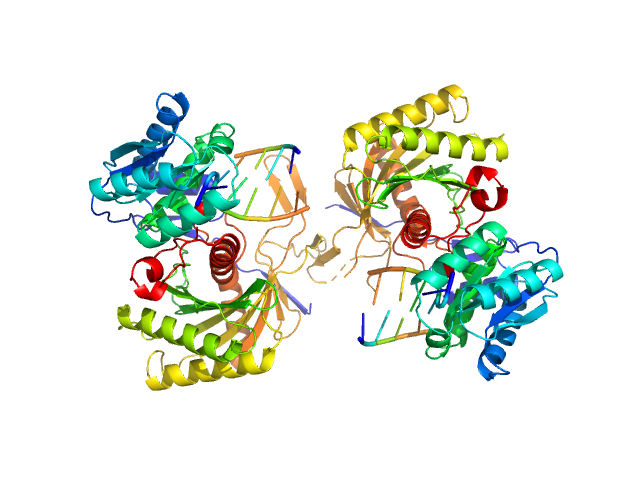
|
| Sample: |
Piwi protein AF_1318 dimer, 102 kDa Archaeoglobus fulgidus protein
5'-phosphorilated 14-mer DNA oligoduplex dimer, 9 kDa DNA
|
| Buffer: |
20 mM TrisHCl, pH 7.5, 5 mM MgCl2, 150 mM NaCl and 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Jul 6
|
Prokaryotic Argonaute from Archaeoglobus fulgidus interacts with DNA as a homodimer.
Sci Rep 11(1):4518 (2021)
Golovinas E, Rutkauskas D, Manakova E, Jankunec M, Silanskas A, Sasnauskas G, Zaremba M
|
| RgGuinier |
3.2 |
nm |
| Dmax |
10.1 |
nm |
| VolumePorod |
158 |
nm3 |
|
|
|
|
|
|
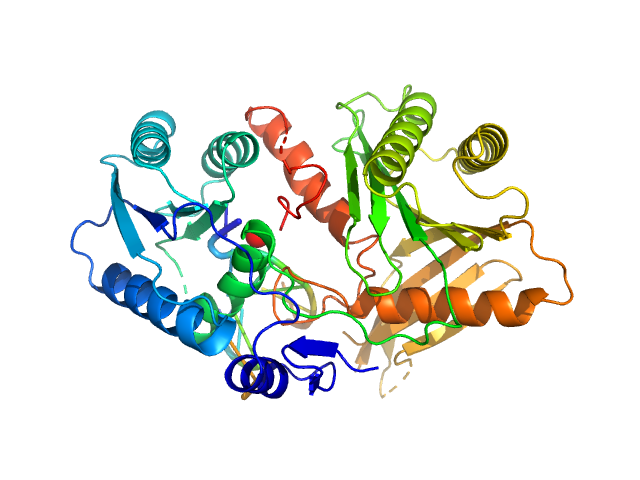
|
| Sample: |
Piwi protein AF_1318 delta (296-303) mutant monomer, 50 kDa Archaeoglobus fulgidus protein
5'-phosphorilated 14-mer DNA oligoduplex dimer, 9 kDa DNA
|
| Buffer: |
20 mM TrisHCl, pH 7.5, 5 mM MgCl2, 150 mM NaCl and 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Jul 6
|
Prokaryotic Argonaute from Archaeoglobus fulgidus interacts with DNA as a homodimer.
Sci Rep 11(1):4518 (2021)
Golovinas E, Rutkauskas D, Manakova E, Jankunec M, Silanskas A, Sasnauskas G, Zaremba M
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.6 |
nm |
| VolumePorod |
109 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nucleolysin TIA-1 isoform p40 monomer, 21 kDa Homo sapiens protein
TC1 monomer, 3 kDa synthetic construct DNA
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 3% v/v glycerol, pH: 7 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2018 Jun 26
|
Tandem RNA binding sites induce self-association of the stress granule marker protein TIA-1.
Nucleic Acids Res (2021)
Loughlin FE, West DL, Gunzburg MJ, Waris S, Crawford SA, Wilce MCJ, Wilce JA
|
| RgGuinier |
3.2 |
nm |
| Dmax |
15.1 |
nm |
| VolumePorod |
78 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nucleolysin TIA-1 isoform p40 monomer, 21 kDa Homo sapiens protein
UC1 monomer, 3 kDa synthetic construct RNA
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 3% v/v glycerol, pH: 7 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2018 Jun 26
|
Tandem RNA binding sites induce self-association of the stress granule marker protein TIA-1.
Nucleic Acids Res (2021)
Loughlin FE, West DL, Gunzburg MJ, Waris S, Crawford SA, Wilce MCJ, Wilce JA
|
| RgGuinier |
3.3 |
nm |
| Dmax |
14.4 |
nm |
| VolumePorod |
77 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
GATA-type iron responsive transcription factor Fep1 reconstituted monomer, 22 kDa Komagataella pastoris protein
24-mer double strand DNA from the GATA promoter dimer, 14 kDa synthetic oligonucleotide DNA
|
| Buffer: |
50 mM MOPS, 50 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Dec 5
|
Biophysical characterization of the complex between the iron-responsive transcription factor Fep1 and DNA.
Eur Biophys J (2021)
Miele AE, Cervoni L, Le Roy A, Cutone A, Musci G, Ebel C, Bonaccorsi di Patti MC
|
| RgGuinier |
3.5 |
nm |
| Dmax |
13.1 |
nm |
| VolumePorod |
73 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
GATA-type iron responsive transcription factor Fep1 monomer, 22 kDa Komagataella pastoris protein
24-mer double strand DNA from the GATA promoter dimer, 14 kDa synthetic oligonucleotide DNA
|
| Buffer: |
50 mM MOPS, 50 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Dec 5
|
Biophysical characterization of the complex between the iron-responsive transcription factor Fep1 and DNA.
Eur Biophys J (2021)
Miele AE, Cervoni L, Le Roy A, Cutone A, Musci G, Ebel C, Bonaccorsi di Patti MC
|
| RgGuinier |
3.2 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
85 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
M100 oligonucleotide monomer, 12 kDa DNA
LIM/homeobox protein Lhx3 monomer, 10 kDa Mus musculus protein
|
| Buffer: |
20 mM sodium phosphate monobasic/dibasic, 100 mM NaCl, 1 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2018 Nov 2
|
Contrasting DNA-binding behaviour by ISL1 and LHX3 underpins differential gene targeting in neuronal cell specification
Journal of Structural Biology: X :100043 (2020)
Smith N, Wilkinson-White L, Kwan A, Trewhella J, Matthews J
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
27 |
nm3 |
|
|
|
|
|
|
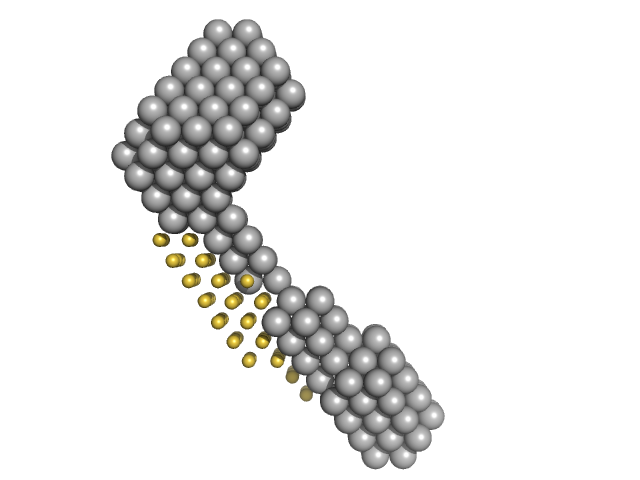
|
| Sample: |
M100 oligonucleotide monomer, 12 kDa DNA
LIM/homeobox protein Lhx3 monomer, 23 kDa Mus musculus protein
Insulin gene enhancer protein ISL-1 monomer, 4 kDa Mus musculus protein
|
| Buffer: |
20 mM sodium phosphate monobasic/dibasic, 100 mM NaCl, 1 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2018 Nov 2
|
Contrasting DNA-binding behaviour by ISL1 and LHX3 underpins differential gene targeting in neuronal cell specification
Journal of Structural Biology: X :100043 (2020)
Smith N, Wilkinson-White L, Kwan A, Trewhella J, Matthews J
|
| RgGuinier |
3.6 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
49 |
nm3 |
|
|
|
|
|
|
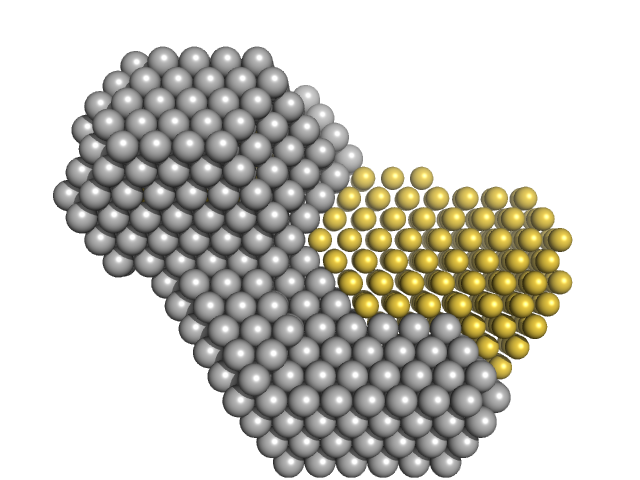
|
| Sample: |
M100 oligonucleotide monomer, 12 kDa DNA
Insulin gene enhancer protein ISL-1 monomer, 12 kDa Mus musculus protein
LIM/homeobox protein Lhx3 monomer, 9 kDa Mus musculus protein
|
| Buffer: |
20 mM sodium phosphate monobasic/dibasic, 100 mM NaCl, 1 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2018 Nov 2
|
Contrasting DNA-binding behaviour by ISL1 and LHX3 underpins differential gene targeting in neuronal cell specification
Journal of Structural Biology: X :100043 (2020)
Smith N, Wilkinson-White L, Kwan A, Trewhella J, Matthews J
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
41 |
nm3 |
|
|
|
|
|
|
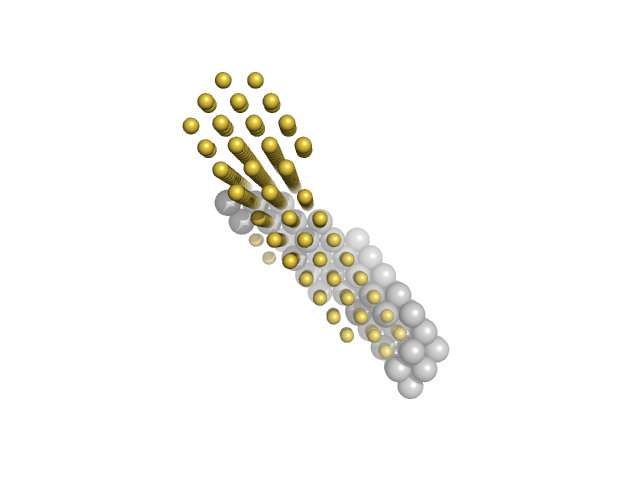
|
| Sample: |
M100 oligonucleotide monomer, 12 kDa DNA
Insulin gene enhancer protein ISL-1 monomer, 14 kDa Mus musculus protein
LIM/homeobox protein Lhx3 monomer, 23 kDa Mus musculus protein
|
| Buffer: |
20 mM sodium phosphate monobasic/dibasic, 100 mM NaCl, 1 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2018 Nov 2
|
Contrasting DNA-binding behaviour by ISL1 and LHX3 underpins differential gene targeting in neuronal cell specification
Journal of Structural Biology: X :100043 (2020)
Smith N, Wilkinson-White L, Kwan A, Trewhella J, Matthews J
|
| RgGuinier |
3.6 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
70 |
nm3 |
|
|