|
|
|
|
|
| Sample: |
Candidatus Glomeribacter gigasporarum cyclodipeptide synthase monomer, 34 kDa Candidatus Glomeribacter gigasporarum protein
E. coli Phe-tRNAPhe monomer, 25 kDa Escherichia coli RNA
|
| Buffer: |
10 mM MOPS pH6.7; 200 mM NaCl, 8 mM MgCl2, pH: 6.7 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2016 Oct 2
|
Structural basis of the interaction between cyclodipeptide synthases and aminoacylated tRNA substrates.
RNA 26(11):1589-1602 (2020)
Bourgeois G, Seguin J, Babin M, Gondry M, Mechulam Y, Schmitt E
|
| RgGuinier |
3.3 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
77 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Primary microRNA pri-miR16-1 complexed with DGCR8-core protein monomer, 36 kDa Homo sapiens RNA
Microprocessor complex subunit DGCR8 monomer, 26 kDa Homo sapiens protein
|
| Buffer: |
50 mM KCl, 50 mM HEPES, 5 mM DTT, 1% glycerol, 50% sucrose, DGCR8-core, pH: 7.5 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2017 Apr 12
|
Elucidating the Role of Microprocessor Protein DGCR8 in Bending RNA Structures
Biophysical Journal (2020)
Pabit S, Chen Y, Usher E, Cook E, Pollack L, Showalter S
|
| RgGuinier |
5.0 |
nm |
| Dmax |
17.2 |
nm |
| VolumePorod |
125 |
nm3 |
|
|
|
|
|
|
|
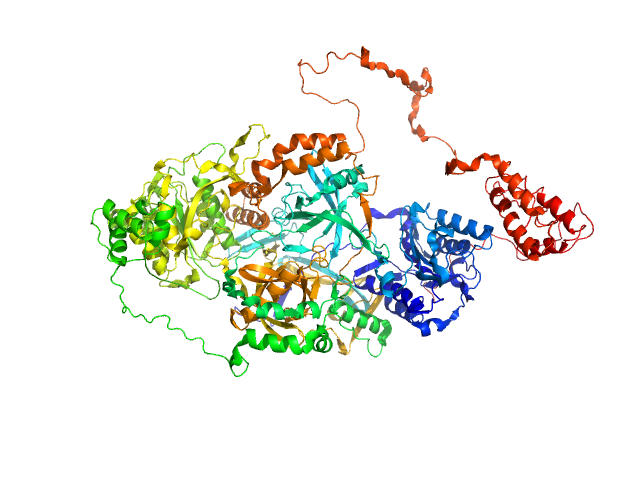
| Sample: |
X-ray repair cross-complementing protein 6 monomer, 70 kDa Homo sapiens protein
X-ray repair cross-complementing protein 5 monomer, 83 kDa Homo sapiens protein
Y-DNA monomer, 18 kDa DNA
|
| Buffer: |
50 mM Tris-HCl, 100 mM NaCl, 5% glycerol, 0.01% sodium azide, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2010 Jan 8
|
Visualizing functional dynamicity in the DNA-dependent protein kinase holoenzyme DNA-PK complex by integrating SAXS with cryo-EM.
Prog Biophys Mol Biol (2020)
Hammel M, Rosenberg DJ, Bierma J, Hura GL, Lees-Miller SP, Tainer JA
|
| RgGuinier |
4.1 |
nm |
| Dmax |
14.8 |
nm |
| VolumePorod |
280 |
nm3 |
|
|
|
|
|
|
|
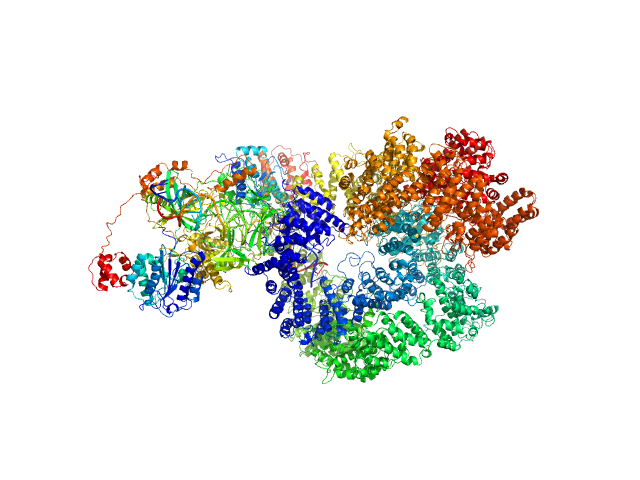
| Sample: |
X-ray repair cross-complementing protein 6 monomer, 70 kDa Homo sapiens protein
X-ray repair cross-complementing protein 5 monomer, 83 kDa Homo sapiens protein
DNA-dependent protein kinase catalytic subunit monomer, 468 kDa Homo sapiens protein
DsDNA dimer, 21 kDa DNA
|
| Buffer: |
50 mM Tris-HCl, 100 mM NaCl, 5% glycerol, 0.01% sodium azide, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Dec 30
|
Visualizing functional dynamicity in the DNA-dependent protein kinase holoenzyme DNA-PK complex by integrating SAXS with cryo-EM.
Prog Biophys Mol Biol (2020)
Hammel M, Rosenberg DJ, Bierma J, Hura GL, Lees-Miller SP, Tainer JA
|
| RgGuinier |
6.5 |
nm |
| Dmax |
23.1 |
nm |
| VolumePorod |
1090 |
nm3 |
|
|
|
|
|
|
|
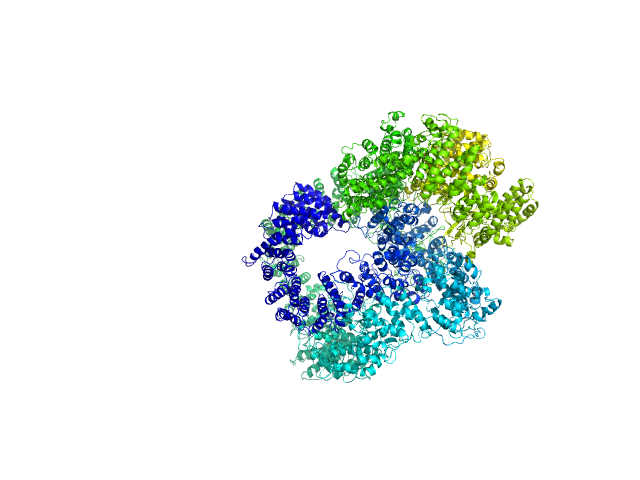
| Sample: |
X-ray repair cross-complementing protein 6 monomer, 70 kDa Homo sapiens protein
X-ray repair cross-complementing protein 5 monomer, 83 kDa Homo sapiens protein
DNA-dependent protein kinase catalytic subunit monomer, 468 kDa Homo sapiens protein
DsDNA dimer, 21 kDa DNA
|
| Buffer: |
50 mM Tris-HCl, 100 mM NaCl, 5% glycerol, 0.01% sodium azide, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Dec 30
|
Visualizing functional dynamicity in the DNA-dependent protein kinase holoenzyme DNA-PK complex by integrating SAXS with cryo-EM.
Prog Biophys Mol Biol (2020)
Hammel M, Rosenberg DJ, Bierma J, Hura GL, Lees-Miller SP, Tainer JA
|
| RgGuinier |
7.5 |
nm |
| Dmax |
29.4 |
nm |
| VolumePorod |
1440 |
nm3 |
|
|
|
|
|
|
|
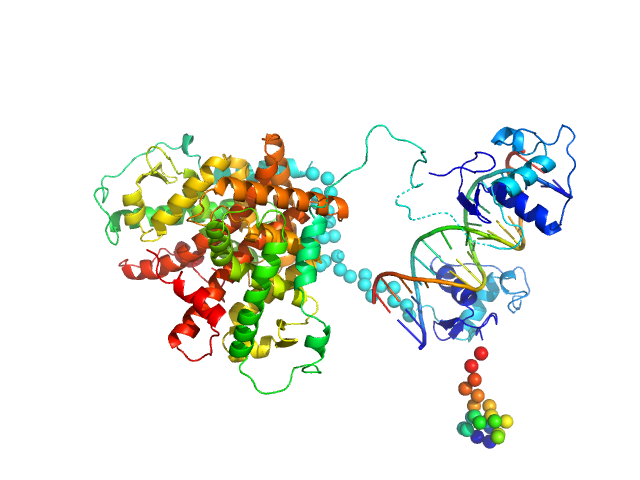
| Sample: |
Retinoic acid receptor alpha, RAR monomer, 41 kDa Mus musculus protein
Retinoic acid receptor RXR-alpha monomer, 38 kDa Mus musculus protein
DNA response element HoxB13 DR0 monomer, 10 kDa DNA
|
| Buffer: |
20 mM Tris, pH 8, 150 mM NaCl, 5% v/v glycerol, 1 mM CHAPS, 4 mM MgSO4, 1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2014 Jan 19
|
Structural basis for DNA recognition and allosteric control of the retinoic acid receptors RAR–RXR
Nucleic Acids Research (2020)
Osz J, McEwen A, Bourguet M, Przybilla F, Peluso-Iltis C, Poussin-Courmontagne P, Mély Y, Cianférani S, Jeffries C, Svergun D, Rochel N
|
| RgGuinier |
3.8 |
nm |
| Dmax |
14.5 |
nm |
| VolumePorod |
132 |
nm3 |
|
|
|
|
|
|
|
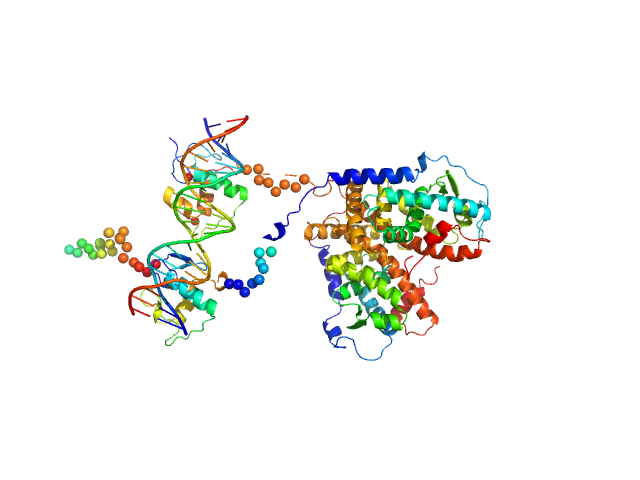
| Sample: |
Retinoic acid receptor alpha, RAR monomer, 41 kDa Mus musculus protein
Retinoic acid receptor RXR-alpha monomer, 38 kDa Mus musculus protein
DNA response element F11r DR5 monomer, 13 kDa DNA
|
| Buffer: |
20 mM Tris, pH 8, 150 mM NaCl, 5% v/v glycerol, 1 mM CHAPS, 4 mM MgSO4, 1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2014 Jan 19
|
Structural basis for DNA recognition and allosteric control of the retinoic acid receptors RAR–RXR
Nucleic Acids Research (2020)
Osz J, McEwen A, Bourguet M, Przybilla F, Peluso-Iltis C, Poussin-Courmontagne P, Mély Y, Cianférani S, Jeffries C, Svergun D, Rochel N
|
| RgGuinier |
4.0 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
130 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Poly(rC)-binding protein 2 monomer, 40 kDa Homo sapiens protein
Modified stem loop IV poliovirus IRES, nucleotides 278-398 monomer, 41 kDa Human poliovirus 1 … RNA
|
| Buffer: |
5 mM HEPES-KOH, 25 mM KCl, 2 mM MgCl2, 2 mM DTT, 4 % glycerol, 0.1 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Dec 16
|
Structure of the PCBP2/stem-loop IV complex underlying translation initiation mediated by the poliovirus type I IRES.
Nucleic Acids Res (2020)
Beckham SA, Matak MY, Belousoff MJ, Venugopal H, Shah N, Vankadari N, Elmlund H, Nguyen JHC, Semler BL, Wilce MCJ, Wilce JA
|
| RgGuinier |
3.7 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
162 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Modified stem loop IV poliovirus IRES, nucleotides 278-398 monomer, 41 kDa Human poliovirus 1 … RNA
Truncated poly(rC)-binding protein 2 (ΔKH3) monomer, 28 kDa Homo sapiens protein
Truncated poly(rC)-binding protein 2 (ΔKH3) monomer, 28 kDa Homo sapiens protein
|
| Buffer: |
5 mM HEPES-KOH, 25 mM KCl, 2 mM MgCl2, 2 mM DTT, 4 % glycerol, 0.1 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Jul 1
|
Structure of the PCBP2/stem-loop IV complex underlying translation initiation mediated by the poliovirus type I IRES.
Nucleic Acids Res (2020)
Beckham SA, Matak MY, Belousoff MJ, Venugopal H, Shah N, Vankadari N, Elmlund H, Nguyen JHC, Semler BL, Wilce MCJ, Wilce JA
|
| RgGuinier |
3.8 |
nm |
| Dmax |
12.2 |
nm |
| VolumePorod |
165 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Modified stem loop IV poliovirus IRES, nucleotides 278-398 monomer, 41 kDa Human poliovirus 1 … RNA
Truncated poly(rC)-binding protein 2 (ΔKH1-KH2) monomer, 18 kDa Homo sapiens protein
|
| Buffer: |
5 mM HEPES-KOH, 25 mM KCl, 2 mM MgCl2, 2 mM DTT, 4 % glycerol, 0.1 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Jul 1
|
Structure of the PCBP2/stem-loop IV complex underlying translation initiation mediated by the poliovirus type I IRES.
Nucleic Acids Res (2020)
Beckham SA, Matak MY, Belousoff MJ, Venugopal H, Shah N, Vankadari N, Elmlund H, Nguyen JHC, Semler BL, Wilce MCJ, Wilce JA
|
| RgGuinier |
3.5 |
nm |
| Dmax |
12.2 |
nm |
| VolumePorod |
126 |
nm3 |
|
|