|
|
|
|
|
| Sample: |
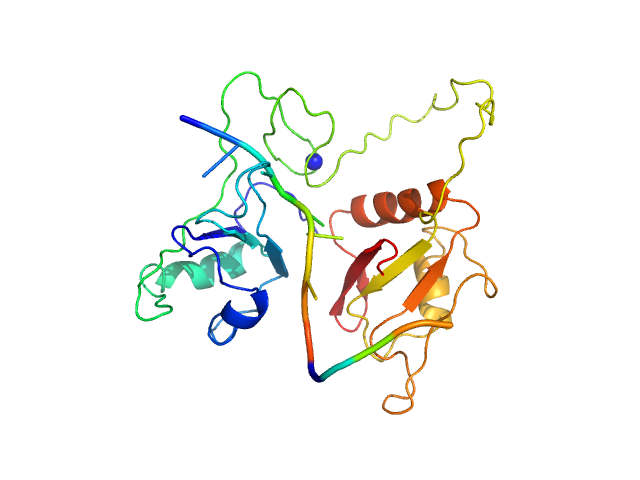
DNA polymerase alpha subunit B monomer, 49 kDa Homo sapiens protein
DNA polymerase alpha catalytic subunit monomer, 23 kDa Homo sapiens protein
DNA primase large subunit monomer, 59 kDa Homo sapiens protein
DNA primase small subunit monomer, 50 kDa Homo sapiens protein
29 mer DNA template monomer, 9 kDa DNA
8mer RNA primer monomer, 3 kDa RNA
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 5 mM MgCl2, 1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2023 Mar 28
|
Flexibility and distributive synthesis regulate RNA priming and handoff in human DNA polymerase α-primase
Journal of Molecular Biology :168330 (2023)
Cordoba J, Mullins E, Salay L, Eichman B, Chazin W
|
| RgGuinier |
4.9 |
nm |
| Dmax |
15.3 |
nm |
| VolumePorod |
344 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
RNA Binding Motif protein 5 (I107T, C191G) monomer, 26 kDa Homo sapiens protein
Caspase-2 derived RNA GGCU_12 monomer, 4 kDa RNA
|
| Buffer: |
20 mM MES, 100 mM NaCl, 1 mM DTT, pH: 6.5 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, SFB 1035, Technische Universität München on 2017 Mar 1
|
Structural basis for specific RNA recognition by the alternative splicing factor RBM5.
Nat Commun 14(1):4233 (2023)
Soni K, Jagtap PKA, Martínez-Lumbreras S, Bonnal S, Geerlof A, Stehle R, Simon B, Valcárcel J, Sattler M
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
42 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nucleoprotein monomer, 15 kDa Severe acute respiratory … protein
Stem loop 2 and 3 in the 5'-genomic end of SARS-CoV-2 monomer, 14 kDa Severe acute respiratory … RNA
|
| Buffer: |
25 mM potassium phosphate, 150 mM KCl, 2 mM TCEP, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Aug 16
|
The preference signature of the SARS-CoV-2 Nucleocapsid NTD for its 5'-genomic RNA elements.
Nat Commun 14(1):3331 (2023)
Korn SM, Dhamotharan K, Jeffries CM, Schlundt A
|
| RgGuinier |
2.4 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
38 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nucleoprotein monomer, 15 kDa Severe acute respiratory … protein
Stem loop 4 with AU extension in the 5'-genomic end of SARS-CoV-2 monomer, 22 kDa Severe acute respiratory … RNA
|
| Buffer: |
25 mM potassium phosphate, 150 mM KCl, 2 mM TCEP, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Aug 16
|
The preference signature of the SARS-CoV-2 Nucleocapsid NTD for its 5'-genomic RNA elements.
Nat Commun 14(1):3331 (2023)
Korn SM, Dhamotharan K, Jeffries CM, Schlundt A
|
| RgGuinier |
3.4 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
51 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nucleoprotein monomer, 15 kDa Severe acute respiratory … protein
AU extension in the 5'-genomic end of SARS-CoV-2 monomer, 7 kDa Severe acute respiratory … RNA
|
| Buffer: |
25 mM potassium phosphate, 150 mM KCl, 2 mM TCEP, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Aug 16
|
The preference signature of the SARS-CoV-2 Nucleocapsid NTD for its 5'-genomic RNA elements.
Nat Commun 14(1):3331 (2023)
Korn SM, Dhamotharan K, Jeffries CM, Schlundt A
|
| RgGuinier |
2.1 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
32 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nucleoprotein monomer, 15 kDa Severe acute respiratory … protein
Stem loop 4 in the 5'-genomic end of SARS-CoV-2 monomer, 14 kDa Severe acute respiratory … RNA
|
| Buffer: |
25 mM potassium phosphate, 150 mM KCl, 2 mM TCEP, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Nov 22
|
The preference signature of the SARS-CoV-2 Nucleocapsid NTD for its 5'-genomic RNA elements.
Nat Commun 14(1):3331 (2023)
Korn SM, Dhamotharan K, Jeffries CM, Schlundt A
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.8 |
nm |
| VolumePorod |
25 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
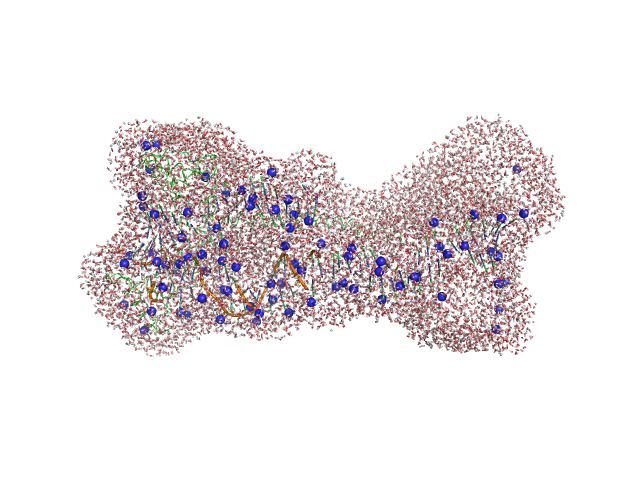
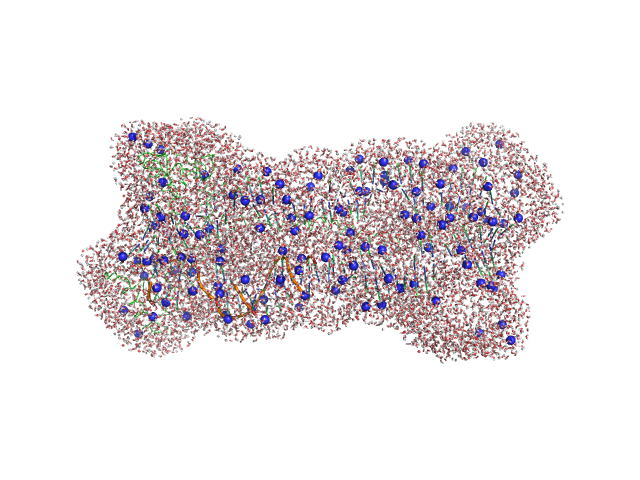
Gcf1p monomer, 26 kDa Candida albicans (strain … protein
Af2_20 DNA monomer, 12 kDa DNA
|
| Buffer: |
25 mM Tris, 20 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Aug 30
|
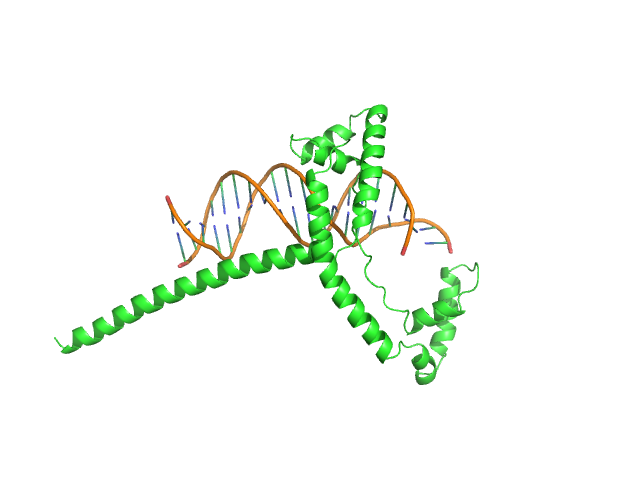
Structural analysis of the Candida albicans mitochondrial DNA maintenance factor Gcf1p reveals a dynamic DNA-bridging mechanism.
Nucleic Acids Res (2023)
Tarrés-Solé A, Battistini F, Gerhold JM, Piétrement O, Martínez-García B, Ruiz-López E, Lyonnais S, Bernadó P, Roca J, Orozco M, Le Cam E, Sedman J, Solà M
|
| RgGuinier |
3.6 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
92 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Gcf1p monomer, 26 kDa Candida albicans (strain … protein
Af2_20 DNA monomer, 12 kDa DNA
|
| Buffer: |
25 mM Tris, 20 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Aug 30
|
Structural analysis of the Candida albicans mitochondrial DNA maintenance factor Gcf1p reveals a dynamic DNA-bridging mechanism.
Nucleic Acids Res (2023)
Tarrés-Solé A, Battistini F, Gerhold JM, Piétrement O, Martínez-García B, Ruiz-López E, Lyonnais S, Bernadó P, Roca J, Orozco M, Le Cam E, Sedman J, Solà M
|
| RgGuinier |
3.9 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
133 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Af2_20 DNA monomer, 12 kDa DNA
Gcf1p(Δ58) monomer, 22 kDa Candida albicans (strain … protein
|
| Buffer: |
25 mM Tris, 20 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Aug 30
|
Structural analysis of the Candida albicans mitochondrial DNA maintenance factor Gcf1p reveals a dynamic DNA-bridging mechanism.
Nucleic Acids Res (2023)
Tarrés-Solé A, Battistini F, Gerhold JM, Piétrement O, Martínez-García B, Ruiz-López E, Lyonnais S, Bernadó P, Roca J, Orozco M, Le Cam E, Sedman J, Solà M
|
| RgGuinier |
2.6 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
52 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
58 nucleotide RNA L11-binding domain from E. coli 23S rRNA monomer, 19 kDa Escherichia coli RNA
50S ribosomal protein L11 monomer, 16 kDa Thermus thermophilus protein
|
| Buffer: |
10 mM Na-MOPSO, 100 mM KCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2018 May 9
|
Chaotic advection mixer for capturing transient states of diverse biological macromolecular systems with time-resolved small-angle X-ray scattering
IUCrJ 10(3):363-375 (2023)
Zielinski K, Katz A, Calvey G, Pabit S, Milano S, Aplin C, San Emeterio J, Cerione R, Pollack L
|
| RgGuinier |
2.4 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
34 |
nm3 |
|
|