|
|
|
|
|
| Sample: |
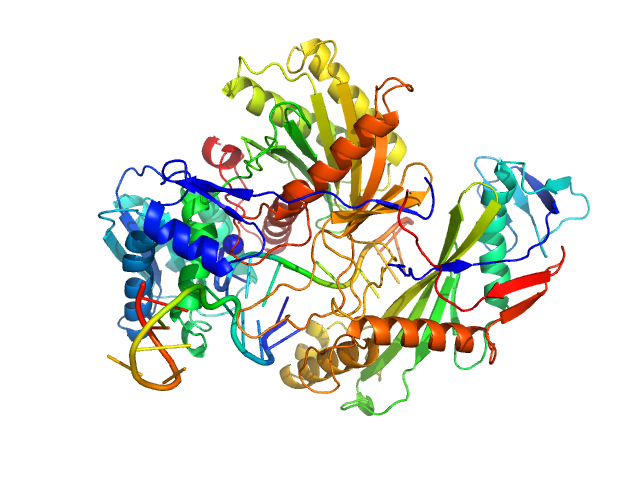
Single-chain full Archaeoglobus fulgidus Argonaute monomer, 78 kDa Archaeoglobus fulgidus protein
5'-end phosphorylated DNA guide strand, 11 nt (MZ864) monomer, 3 kDa DNA
DNA target strand, 11 nt (MZ865) monomer, 3 kDa DNA
|
| Buffer: |
20 mM TrisHCl pH7.5, 200 mM NaCl, 5 mM MgCl2, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Aug 31
|
The missing part: the Archaeoglobus fulgidus Argonaute forms a functional heterodimer with an N-L1-L2 domain protein.
Nucleic Acids Res (2024)
Manakova E, Golovinas E, Pocevičiūtė R, Sasnauskas G, Silanskas A, Rutkauskas D, Jankunec M, Zagorskaitė E, Jurgelaitis E, Grybauskas A, Venclovas Č, Zaremba M
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.6 |
nm |
| VolumePorod |
125 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
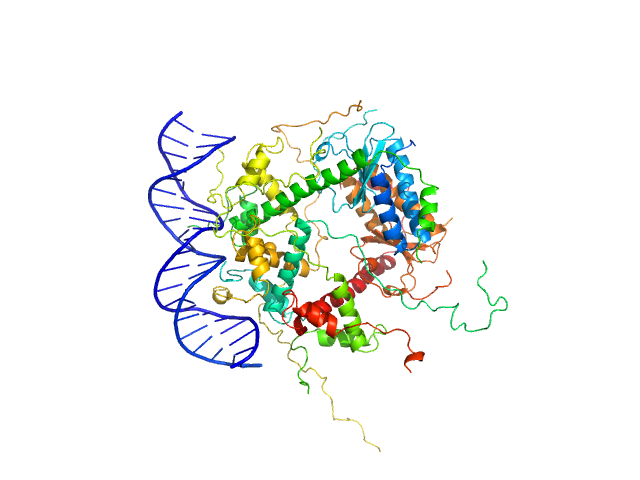
Antitoxin ParD hexamer, 54 kDa Vibrio cholerae serotype … protein
Toxin, 25 kDa Vibrio cholerae serotype … protein
21-bp DNA operator fragment monomer, 13 kDa Vibrio cholerae O1 DNA
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2020 Jul 18
|
Toxin:antitoxin ratio sensing autoregulation of the Vibrio cholerae parDE2 module.
Sci Adv 10(1):eadj2403 (2024)
Garcia-Rodriguez G, Girardin Y, Kumar Singh R, Volkov AN, Van Dyck J, Muruganandam G, Sobott F, Charlier D, Loris R
|
| RgGuinier |
3.2 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
140 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
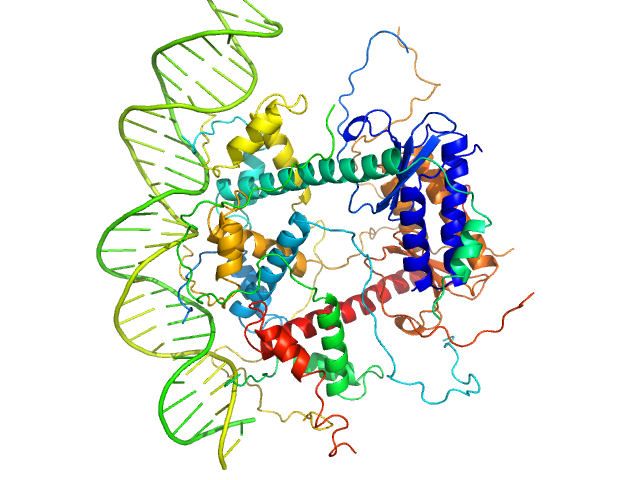
Antitoxin ParD hexamer, 54 kDa Vibrio cholerae serotype … protein
Toxin, 25 kDa Vibrio cholerae serotype … protein
31-bp DNA operator box monomer, 19 kDa Vibrio cholerae O1 DNA
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Dec 4
|
Toxin:antitoxin ratio sensing autoregulation of the Vibrio cholerae parDE2 module.
Sci Adv 10(1):eadj2403 (2024)
Garcia-Rodriguez G, Girardin Y, Kumar Singh R, Volkov AN, Van Dyck J, Muruganandam G, Sobott F, Charlier D, Loris R
|
| RgGuinier |
3.3 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
160 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
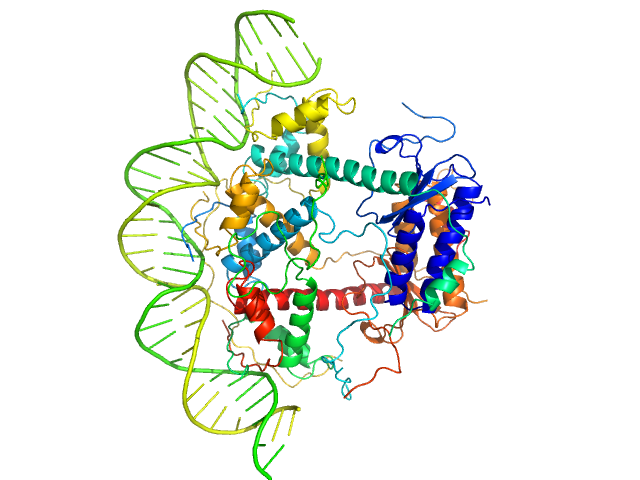
Antitoxin ParD hexamer, 54 kDa Vibrio cholerae serotype … protein
Toxin, 25 kDa Vibrio cholerae serotype … protein
33-bp DNA operator fragment monomer, 20 kDa Vibrio cholerae O1 DNA
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2020 Jul 18
|
Toxin:antitoxin ratio sensing autoregulation of the Vibrio cholerae parDE2 module.
Sci Adv 10(1):eadj2403 (2024)
Garcia-Rodriguez G, Girardin Y, Kumar Singh R, Volkov AN, Van Dyck J, Muruganandam G, Sobott F, Charlier D, Loris R
|
| RgGuinier |
3.2 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
150 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Queuine tRNA-ribosyltransferase catalytic subunit 1 monomer, 44 kDa Homo sapiens protein
Queuine tRNA-ribosyltransferase accessory subunit 2 monomer, 47 kDa Homo sapiens protein
Transfer RNA (Aspartate) monomer, 25 kDa Homo sapiens RNA
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 3% (w/v) glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Dec 1
|
Structural and functional investigation of tRNA guanine transglycosylase
University of Göttingen Dissertation - (2023)
Katharina Sievers
|
| RgGuinier |
3.5 |
nm |
| Dmax |
10.9 |
nm |
| VolumePorod |
160 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Stem loop 2 and 3 in the 5'-genomic end of SARS-CoV-2 monomer, 14 kDa Severe acute respiratory … RNA
Nucleoprotein dimer, 30 kDa Severe acute respiratory … protein
|
| Buffer: |
25 mM HEPES, 75 mM KCl, 2.5 mM NaNO3, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Nov 29
|
The preference signature of the SARS-CoV-2 Nucleocapsid NTD for its 5'-genomic RNA elements
Karthikeyan Dhamotharan
|
| RgGuinier |
2.9 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
69 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nucleoprotein monomer, 15 kDa Severe acute respiratory … protein
Stem loop 4 with AU extension in the 5'-genomic end of SARS-CoV-2 monomer, 22 kDa Severe acute respiratory … RNA
|
| Buffer: |
25 mM HEPES, 75 mM KCl, 2.5 mM NaNO3, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Nov 29
|
The preference signature of the SARS-CoV-2 Nucleocapsid NTD for its 5'-genomic RNA elements
Karthikeyan Dhamotharan
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.8 |
nm |
| VolumePorod |
50 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
AU extension in the 5'-genomic end of SARS-CoV-2 monomer, 7 kDa Severe acute respiratory … RNA
Nucleoprotein dimer, 30 kDa Severe acute respiratory … protein
|
| Buffer: |
25 mM HEPES, 75 mM KCl, 2.5 mM NaNO3, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Nov 29
|
The preference signature of the SARS-CoV-2 Nucleocapsid NTD for its 5'-genomic RNA elements
Karthikeyan Dhamotharan
|
| RgGuinier |
2.6 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
46 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nucleoprotein monomer, 15 kDa Severe acute respiratory … protein
Stem loop 4 in the 5'-genomic end of SARS-CoV-2 monomer, 14 kDa Severe acute respiratory … RNA
|
| Buffer: |
25 mM HEPES, 75 mM KCl, 2.5 mM NaNO3, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Nov 29
|
The preference signature of the SARS-CoV-2 Nucleocapsid NTD for its 5'-genomic RNA elements
Karthikeyan Dhamotharan
|
| RgGuinier |
2.7 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
46 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
DNA polymerase alpha subunit B monomer, 49 kDa Homo sapiens protein
DNA polymerase alpha catalytic subunit monomer, 23 kDa Homo sapiens protein
DNA primase large subunit monomer, 59 kDa Homo sapiens protein
DNA primase small subunit monomer, 50 kDa Homo sapiens protein
22mer DNA template monomer, 7 kDa DNA
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 5 mM MnCl2, 1 mM TCEP, 1 mM 5'-guanylylmethylenediphosphonate (GMPCPP), pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Dec 4
|
Flexibility and distributive synthesis regulate RNA priming and handoff in human DNA polymerase α-primase
Journal of Molecular Biology :168330 (2023)
Cordoba J, Mullins E, Salay L, Eichman B, Chazin W
|
| RgGuinier |
4.6 |
nm |
| Dmax |
14.2 |
nm |
| VolumePorod |
310 |
nm3 |
|
|