|
|
|
|
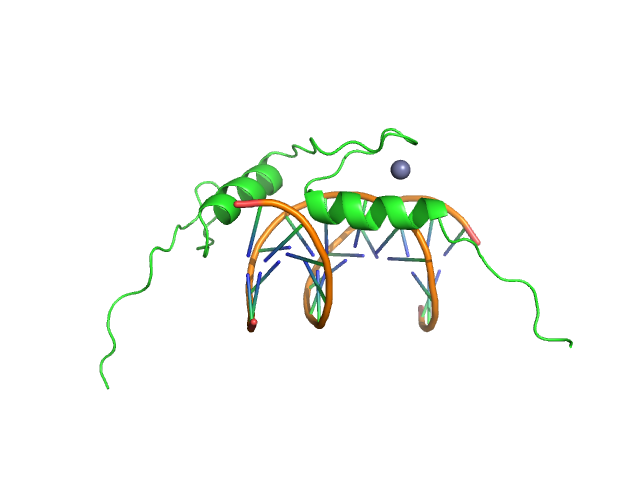
|
| Sample: |
Sal-like protein 4 monomer, 8 kDa Mus musculus protein
AT-rich dsDNA monomer, 7 kDa DNA
|
| Buffer: |
20 mM Tris-HCl, pH 7.5, 200 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2020 Feb 6
|
Structure of SALL4 zinc finger domain reveals link between AT-rich DNA binding and Okihiro syndrome.
Life Sci Alliance 6(3) (2023)
Watson JA, Pantier R, Jayachandran U, Chhatbar K, Alexander-Howden B, Kruusvee V, Prendecki M, Bird A, Cook AG
|
| RgGuinier |
1.8 |
nm |
| Dmax |
5.1 |
nm |
| VolumePorod |
14 |
nm3 |
|
|
|
|
|
|
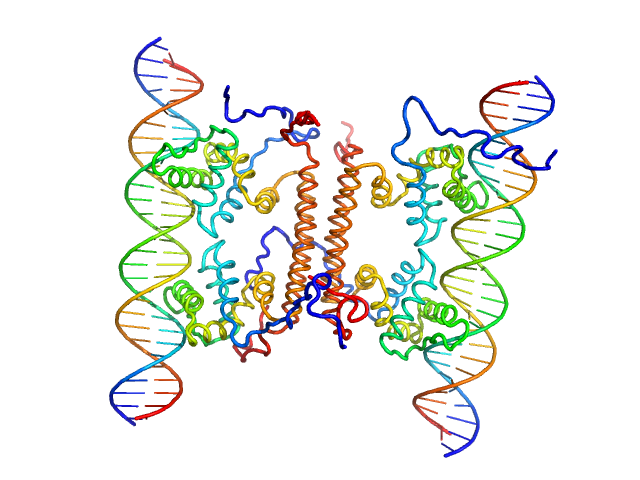
|
| Sample: |
YdaT_toxin domain-containing protein tetramer, 74 kDa Escherichia coli O157:H7 protein
Om 30 base pair dsDNA dimer, 37 kDa DNA
|
| Buffer: |
20 mM Tris-HCl, 200 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Apr 14
|
Structural basis of DNA binding by YdaT, a functional equivalent of the CII repressor in the cryptic prophage CP-933P from Escherichia coli
O157:H7
Acta Crystallographica Section D Structural Biology 79(3):245-258 (2023)
Prolič-Kalinšek M, Volkov A, Hadži S, Van Dyck J, Bervoets I, Charlier D, Loris R
|
| RgGuinier |
4.2 |
nm |
| Dmax |
12.8 |
nm |
| VolumePorod |
190 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Eukaryotic initiation factor 4A-I monomer, 46 kDa Homo sapiens protein
(AG)10-RNA monomer, 7 kDa synthetic construct RNA
|
| Buffer: |
20 mM Hepes, 100 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Sep 16
|
eIF4A1-dependent mRNAs employ purine-rich 5'UTR sequences to activate localised eIF4A1-unwinding through eIF4A1-multimerisation to facilitate translation.
Nucleic Acids Res (2023)
Schmidt T, Dabrowska A, Waldron JA, Hodge K, Koulouras G, Gabrielsen M, Munro J, Tack DC, Harris G, McGhee E, Scott D, Carlin LM, Huang D, Le Quesne J, Zanivan S, Wilczynska A, Bushell M
|
| RgGuinier |
2.8 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
96 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Eukaryotic initiation factor 4A-I monomer, 46 kDa Homo sapiens protein
(CAA)6CA-RNA monomer, 6 kDa synthetic construct RNA
|
| Buffer: |
20 mM Hepes, 100 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Jul 22
|
eIF4A1-dependent mRNAs employ purine-rich 5'UTR sequences to activate localised eIF4A1-unwinding through eIF4A1-multimerisation to facilitate translation.
Nucleic Acids Res (2023)
Schmidt T, Dabrowska A, Waldron JA, Hodge K, Koulouras G, Gabrielsen M, Munro J, Tack DC, Harris G, McGhee E, Scott D, Carlin LM, Huang D, Le Quesne J, Zanivan S, Wilczynska A, Bushell M
|
| RgGuinier |
3.1 |
nm |
| Dmax |
12.8 |
nm |
| VolumePorod |
89 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
(AG)10-RNA monomer, 7 kDa synthetic construct RNA
Eukaryotic initiation factor 4A-I trimer, 138 kDa Homo sapiens protein
|
| Buffer: |
20 mM Hepes, 100 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Sep 16
|
eIF4A1-dependent mRNAs employ purine-rich 5'UTR sequences to activate localised eIF4A1-unwinding through eIF4A1-multimerisation to facilitate translation.
Nucleic Acids Res (2023)
Schmidt T, Dabrowska A, Waldron JA, Hodge K, Koulouras G, Gabrielsen M, Munro J, Tack DC, Harris G, McGhee E, Scott D, Carlin LM, Huang D, Le Quesne J, Zanivan S, Wilczynska A, Bushell M
|
| RgGuinier |
3.4 |
nm |
| Dmax |
13.1 |
nm |
| VolumePorod |
150 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Eukaryotic initiation factor 4A-I trimer, 138 kDa Homo sapiens protein
(AG)10-24bp loading-RNA monomer, 15 kDa synthetic construct RNA
(AG)10-24bp duplex-RNA monomer, 7 kDa synthetic construct RNA
|
| Buffer: |
20 mM Hepes, 100 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Sep 16
|
eIF4A1-dependent mRNAs employ purine-rich 5'UTR sequences to activate localised eIF4A1-unwinding through eIF4A1-multimerisation to facilitate translation.
Nucleic Acids Res (2023)
Schmidt T, Dabrowska A, Waldron JA, Hodge K, Koulouras G, Gabrielsen M, Munro J, Tack DC, Harris G, McGhee E, Scott D, Carlin LM, Huang D, Le Quesne J, Zanivan S, Wilczynska A, Bushell M
|
| RgGuinier |
4.9 |
nm |
| Dmax |
16.2 |
nm |
| VolumePorod |
230 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Upstream of N-ras, isoform A monomer, 26 kDa Drosophila melanogaster protein
PolyA-15mer monomer, 5 kDa RNA
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 8
|
Upstream of N-Ras C-terminal cold shock domains mediate poly(A) specificity in a novel RNA recognition mode and bind poly(A) binding protein.
Nucleic Acids Res (2023)
Hollmann NM, Jagtap PKA, Linse JB, Ullmann P, Payr M, Murciano B, Simon B, Hub JS, Hennig J
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
37 |
nm3 |
|
|
|
|
|
|
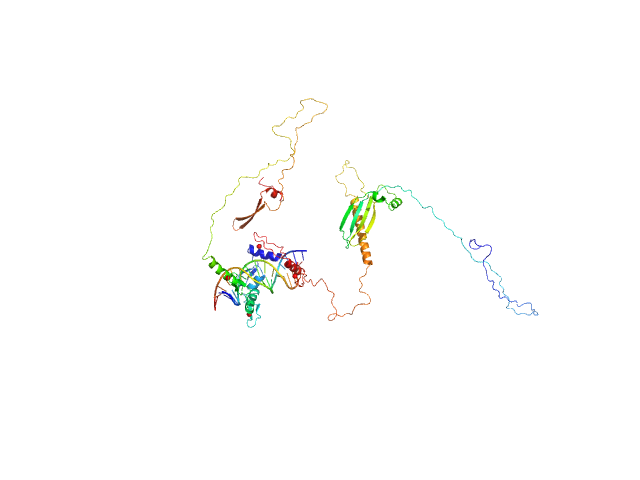
|
| Sample: |
Zinc finger protein 410 monomer, 52 kDa Homo sapiens protein
DNA (Zinc finger protein 410 recognition sequence) monomer, 11 kDa DNA
|
| Buffer: |
20 mM Tris, 250 mM NaCl, 0.1% v/v β-mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Sep 30
|
Allosteric autoregulation of DNA binding via a DNA-mimicking protein domain: a biophysical study of ZNF410-DNA interaction using small angle X-ray scattering.
Nucleic Acids Res (2023)
Kaur G, Ren R, Hammel M, Horton JR, Yang J, Cao Y, He C, Lan F, Lan X, Blobel GA, Blumenthal RM, Zhang X, Cheng X
|
| RgGuinier |
4.4 |
nm |
| Dmax |
14.3 |
nm |
| VolumePorod |
76 |
nm3 |
|
|
|
|
|
|
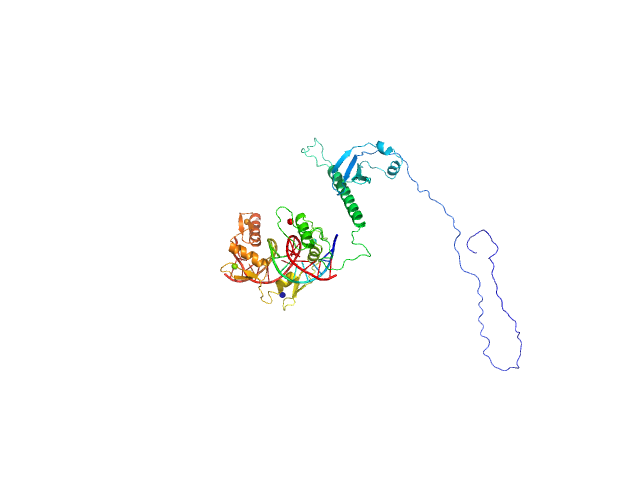
|
| Sample: |
DNA (Zinc finger protein 410 recognition sequence) monomer, 11 kDa DNA
Zinc finger protein 410 monomer, 40 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 250 mM NaCl, 0.1% v/v β-mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2021 Apr 25
|
Allosteric autoregulation of DNA binding via a DNA-mimicking protein domain: a biophysical study of ZNF410-DNA interaction using small angle X-ray scattering.
Nucleic Acids Res (2023)
Kaur G, Ren R, Hammel M, Horton JR, Yang J, Cao Y, He C, Lan F, Lan X, Blobel GA, Blumenthal RM, Zhang X, Cheng X
|
| RgGuinier |
3.1 |
nm |
| Dmax |
14.1 |
nm |
| VolumePorod |
63 |
nm3 |
|
|
|
|
|
|
|
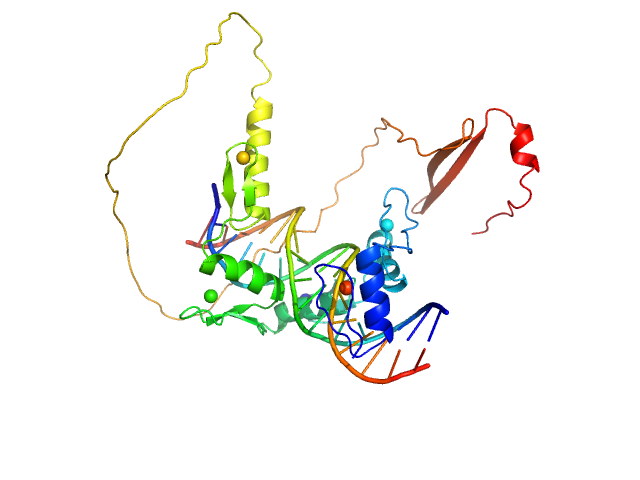
| Sample: |
DNA (Zinc finger protein 410 recognition sequence) monomer, 11 kDa DNA
Zinc finger protein 410 monomer, 29 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 250 mM NaCl, 0.1% v/v β-mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Sep 30
|
Allosteric autoregulation of DNA binding via a DNA-mimicking protein domain: a biophysical study of ZNF410-DNA interaction using small angle X-ray scattering.
Nucleic Acids Res (2023)
Kaur G, Ren R, Hammel M, Horton JR, Yang J, Cao Y, He C, Lan F, Lan X, Blobel GA, Blumenthal RM, Zhang X, Cheng X
|
| RgGuinier |
3.1 |
nm |
| Dmax |
11.9 |
nm |
| VolumePorod |
52 |
nm3 |
|
|