|
|
|
|
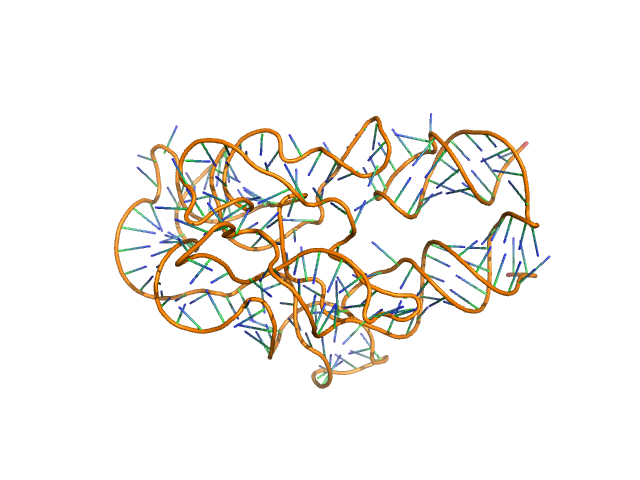
|
| Sample: |
Thermoanearobacter tengcongensis (Tte) fecB riboswitch aptamer domain, 68 kDa marine metagenome RNA
|
| Buffer: |
50 mM MES pH 6.0, 10 mM KCl, 1 mM MgCl2, pH: 6 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2021 Apr 18
|
Visualizing RNA conformational and architectural heterogeneity in solution.
Nat Commun 14(1):714 (2023)
Ding J, Lee YT, Bhandari Y, Schwieters CD, Fan L, Yu P, Tarosov SG, Stagno JR, Ma B, Nussinov R, Rein A, Zhang J, Wang YX
|
| RgGuinier |
5.9 |
nm |
| Dmax |
20.7 |
nm |
| VolumePorod |
206 |
nm3 |
|
|
|
|
|
|
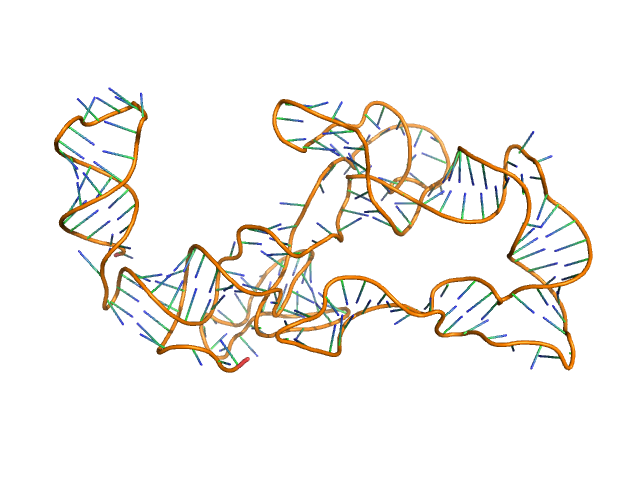
|
| Sample: |
Thermoanearobacter tengcongensis (Tte) fecB riboswitch aptamer domain, 68 kDa marine metagenome RNA
|
| Buffer: |
50 mM MES pH 6.0, 10 mM KCl, 1 mM MgCl2, 4-18 μM coenzyme B12 ligand, pH: 6 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2021 Apr 18
|
Visualizing RNA conformational and architectural heterogeneity in solution.
Nat Commun 14(1):714 (2023)
Ding J, Lee YT, Bhandari Y, Schwieters CD, Fan L, Yu P, Tarosov SG, Stagno JR, Ma B, Nussinov R, Rein A, Zhang J, Wang YX
|
| RgGuinier |
5.7 |
nm |
| Dmax |
19.7 |
nm |
| VolumePorod |
230 |
nm3 |
|
|
|
|
|
|
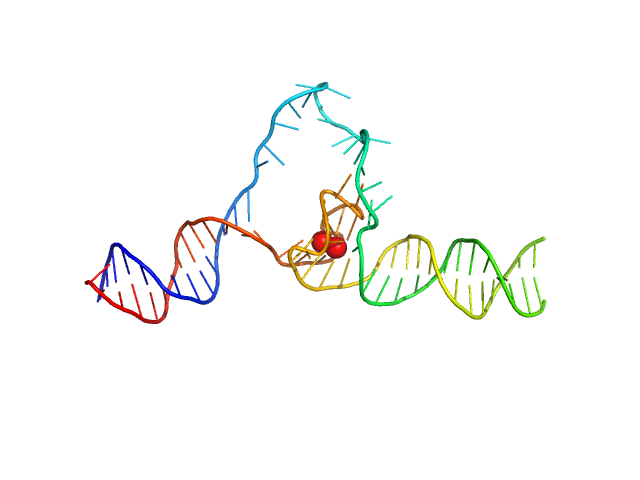
|
| Sample: |
Duplex-G4-duplex monomer, 29 kDa synthetic construct DNA
|
| Buffer: |
6 mM Na2HPO4, 2 mM NaH2PO4, 1 mM Na2EDTA, 185 mM KCl, pH: 7.2 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2021 Jun 16
|
Structure of a 28.5 kDa duplex-embedded G-quadruplex system resolved to 7.4 Å resolution with cryo-EM.
Nucleic Acids Res (2023)
Monsen RC, Chua EYD, Hopkins JB, Chaires JB, Trent JO
|
| RgGuinier |
3.2 |
nm |
| Dmax |
13.3 |
nm |
| VolumePorod |
59 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
DNA (Zinc finger protein 410 recognition sequence) monomer, 11 kDa DNA
|
| Buffer: |
20 mM Tris, 250 mM NaCl, 0.1% v/v β-mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Sep 30
|
Allosteric autoregulation of DNA binding via a DNA-mimicking protein domain: a biophysical study of ZNF410-DNA interaction using small angle X-ray scattering.
Nucleic Acids Res (2023)
Kaur G, Ren R, Hammel M, Horton JR, Yang J, Cao Y, He C, Lan F, Lan X, Blobel GA, Blumenthal RM, Zhang X, Cheng X
|
| RgGuinier |
1.8 |
nm |
| Dmax |
5.8 |
nm |
| VolumePorod |
16 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Alpha cyclodextrin monomer, 1 kDa
|
| Buffer: |
water, pH: 7 |
| Experiment: |
SAXS
data collected at 11-ID-B, Advanced Photon Source (APS), Argonne National Laboratory on 2021 Nov 2
|
Extended q‐range X‐ray Scattering Reveals High‐Resolution Structural Details of Biomacromolecules in Aqueous Solutions
Chemistry – A European Journal (2023)
Chen J, Borkiewicz O, Grishaev A, Zhang F, Bera M, Ruett U, Levin I
|
|
|
|
|
|
|
|
| Sample: |
Alpha cyclodextrin monomer, 1 kDa
|
| Buffer: |
water, pH: 7 |
| Experiment: |
SAXS
data collected at 11-ID-B, Advanced Photon Source (APS), Argonne National Laboratory on 2021 Nov 2
|
Extended q‐range X‐ray Scattering Reveals High‐Resolution Structural Details of Biomacromolecules in Aqueous Solutions
Chemistry – A European Journal (2023)
Chen J, Borkiewicz O, Grishaev A, Zhang F, Bera M, Ruett U, Levin I
|
|
|
|
|
|
|
|
| Sample: |
Alpha cyclodextrin monomer, 1 kDa
|
| Buffer: |
water, pH: 7 |
| Experiment: |
SAXS
data collected at 11-ID-B, Advanced Photon Source (APS), Argonne National Laboratory on 2021 Nov 2
|
Extended q‐range X‐ray Scattering Reveals High‐Resolution Structural Details of Biomacromolecules in Aqueous Solutions
Chemistry – A European Journal (2023)
Chen J, Borkiewicz O, Grishaev A, Zhang F, Bera M, Ruett U, Levin I
|
|
|
|
|
|
|
|
| Sample: |
Beta Cyclodextrin monomer, 1 kDa
|
| Buffer: |
water, pH: 7 |
| Experiment: |
SAXS
data collected at 11-ID-B, Advanced Photon Source (APS), Argonne National Laboratory on 2021 Nov 2
|
Extended q‐range X‐ray Scattering Reveals High‐Resolution Structural Details of Biomacromolecules in Aqueous Solutions
Chemistry – A European Journal (2023)
Chen J, Borkiewicz O, Grishaev A, Zhang F, Bera M, Ruett U, Levin I
|
|
|
|
|
|
|
|
| Sample: |
Beta Cyclodextrin monomer, 1 kDa
|
| Buffer: |
water, pH: 7 |
| Experiment: |
SAXS
data collected at 11-ID-B, Advanced Photon Source (APS), Argonne National Laboratory on 2021 Nov 2
|
Extended q‐range X‐ray Scattering Reveals High‐Resolution Structural Details of Biomacromolecules in Aqueous Solutions
Chemistry – A European Journal (2023)
Chen J, Borkiewicz O, Grishaev A, Zhang F, Bera M, Ruett U, Levin I
|
|
|
|
|
|
|
|
| Sample: |
Gamma Cyclodextrin monomer, 1 kDa
|
| Buffer: |
water, pH: 7 |
| Experiment: |
SAXS
data collected at 11-ID-B, Advanced Photon Source (APS), Argonne National Laboratory on 2021 Nov 2
|
Extended q‐range X‐ray Scattering Reveals High‐Resolution Structural Details of Biomacromolecules in Aqueous Solutions
Chemistry – A European Journal (2023)
Chen J, Borkiewicz O, Grishaev A, Zhang F, Bera M, Ruett U, Levin I
|
|
|