|
|
|
|

|
| Sample: |
Zinc protease PqqL monomer, 102 kDa Escherichia coli protein
|
| Buffer: |
20 mM Tris HCl, 150 nM NaCl, 0.02 % NaN3, 5% glycerol, pH: 7.8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Jun 14
|
Protease-associated import systems are widespread in Gram-negative bacteria.
PLoS Genet 15(10):e1008435 (2019)
Grinter R, Leung PM, Wijeyewickrema LC, Littler D, Beckham S, Pike RN, Walker D, Greening C, Lithgow T
|
| RgGuinier |
4.0 |
nm |
| Dmax |
13.7 |
nm |
| VolumePorod |
207 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
4-hydroxy-tetrahydrodipicolinate synthase tetramer, 131 kDa Campylobacter jejuni protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Aug 2
|
Asparagine-84, a regulatory allosteric site residue, helps maintain the quaternary structure of Campylobacter jejuni dihydrodipicolinate synthase.
J Struct Biol :107409 (2019)
Majdi Yazdi M, Saran S, Mrozowich T, Lehnert C, Patel TR, Sanders DAR, Palmer DRJ
|
| RgGuinier |
3.4 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
188 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
4-hydroxy-tetrahydrodipicolinate synthase (N84D mutant) dimer, 65 kDa Campylobacter jejuni protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Aug 2
|
Asparagine-84, a regulatory allosteric site residue, helps maintain the quaternary structure of Campylobacter jejuni dihydrodipicolinate synthase.
J Struct Biol :107409 (2019)
Majdi Yazdi M, Saran S, Mrozowich T, Lehnert C, Patel TR, Sanders DAR, Palmer DRJ
|
| RgGuinier |
3.1 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
108 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
4-hydroxy-tetrahydrodipicolinate synthase (N84A mutant), 33 kDa Campylobacter jejuni protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Aug 2
|
Asparagine-84, a regulatory allosteric site residue, helps maintain the quaternary structure of Campylobacter jejuni dihydrodipicolinate synthase.
J Struct Biol :107409 (2019)
Majdi Yazdi M, Saran S, Mrozowich T, Lehnert C, Patel TR, Sanders DAR, Palmer DRJ
|
| RgGuinier |
3.3 |
nm |
| Dmax |
8.9 |
nm |
| VolumePorod |
163 |
nm3 |
|
|
|
|
|
|
|
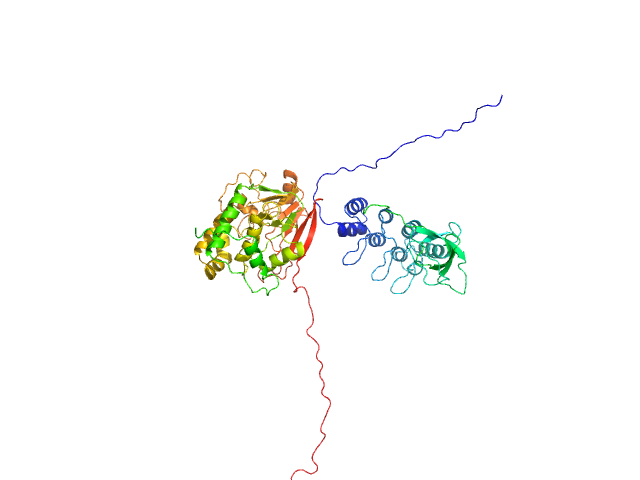
| Sample: |
Serine/threonine-protein phosphatase PP1-alpha catalytic subunit monomer, 38 kDa Homo sapiens protein
Inhibitor of apoptosis-stimulating protein of p53 (RelA-associated inhibitor) monomer, 25 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris, 150 mM NaCl, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Apr 25
|
Flexible Tethering of ASPP Proteins Facilitates PP-1c Catalysis.
Structure 27(10):1485-1496.e4 (2019)
Zhou Y, Millott R, Kim HJ, Peng S, Edwards RA, Skene-Arnold T, Hammel M, Lees-Miller SP, Tainer JA, Holmes CFB, Glover JNM
|
| RgGuinier |
3.4 |
nm |
| Dmax |
12.4 |
nm |
| VolumePorod |
132 |
nm3 |
|
|
|
|
|
|
|
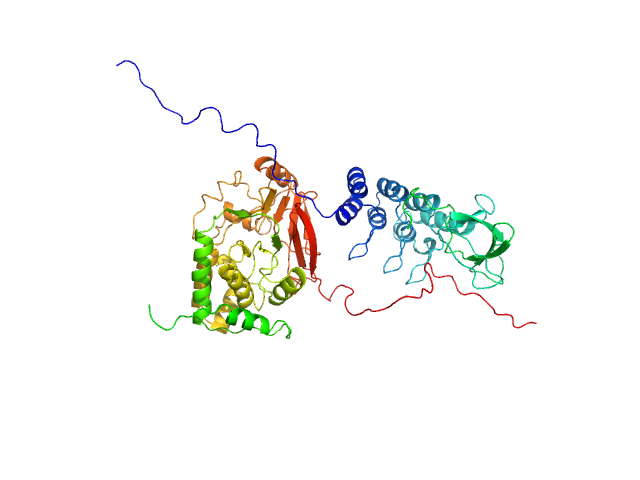
| Sample: |
Serine/threonine-protein phosphatase PP1-alpha catalytic subunit monomer, 38 kDa Homo sapiens protein
Inhibitor of apoptosis-stimulating protein of p53 (RelA-associated inhibitor) monomer, 25 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris, 150 mM NaCl, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Apr 25
|
Flexible Tethering of ASPP Proteins Facilitates PP-1c Catalysis.
Structure 27(10):1485-1496.e4 (2019)
Zhou Y, Millott R, Kim HJ, Peng S, Edwards RA, Skene-Arnold T, Hammel M, Lees-Miller SP, Tainer JA, Holmes CFB, Glover JNM
|
| RgGuinier |
3.0 |
nm |
| Dmax |
10.9 |
nm |
| VolumePorod |
116 |
nm3 |
|
|
|
|
|
|
|
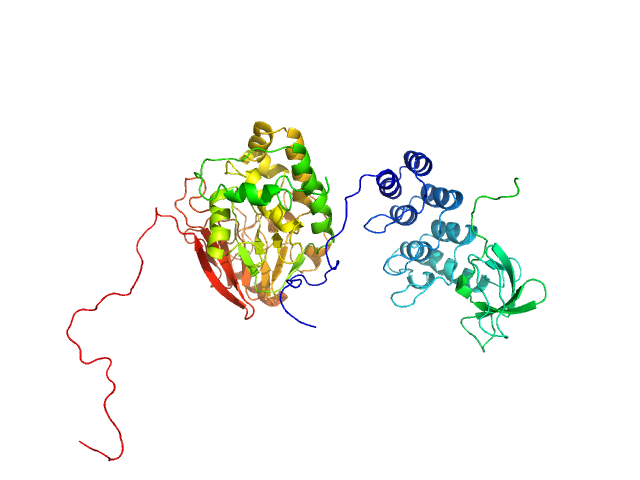
| Sample: |
Inhibitor of apoptosis-stimulating protein of p53 (RelA-associated inhibitor) monomer, 25 kDa Homo sapiens protein
Serine/threonine-protein phosphatase PP1-alpha catalytic subunit monomer, 38 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris, 150 mM NaCl, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 May 31
|
Flexible Tethering of ASPP Proteins Facilitates PP-1c Catalysis.
Structure 27(10):1485-1496.e4 (2019)
Zhou Y, Millott R, Kim HJ, Peng S, Edwards RA, Skene-Arnold T, Hammel M, Lees-Miller SP, Tainer JA, Holmes CFB, Glover JNM
|
| RgGuinier |
3.1 |
nm |
| Dmax |
11.3 |
nm |
| VolumePorod |
157 |
nm3 |
|
|
|
|
|
|
|
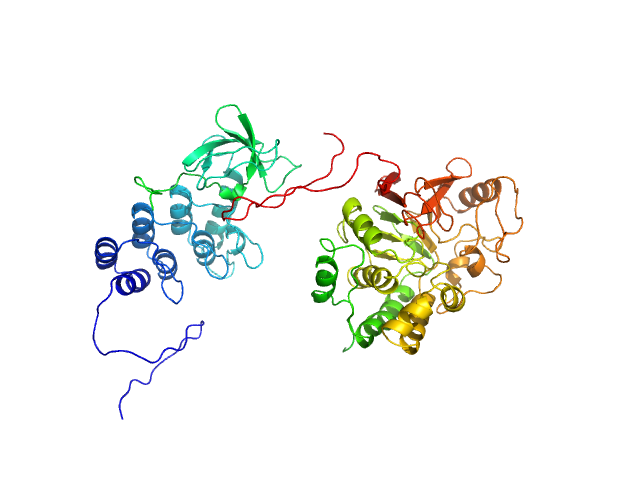
| Sample: |
Inhibitor of apoptosis-stimulating protein of p53 (RelA-associated inhibitor) monomer, 25 kDa Homo sapiens protein
Serine/threonine-protein phosphatase PP1-alpha catalytic subunit monomer, 38 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris, 150 mM NaCl, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 May 31
|
Flexible Tethering of ASPP Proteins Facilitates PP-1c Catalysis.
Structure 27(10):1485-1496.e4 (2019)
Zhou Y, Millott R, Kim HJ, Peng S, Edwards RA, Skene-Arnold T, Hammel M, Lees-Miller SP, Tainer JA, Holmes CFB, Glover JNM
|
| RgGuinier |
3.4 |
nm |
| Dmax |
12.2 |
nm |
| VolumePorod |
180 |
nm3 |
|
|
|
|
|
|
|
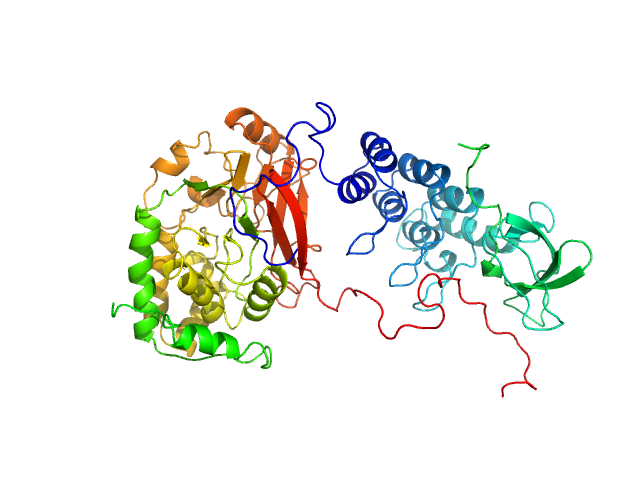
| Sample: |
Serine/threonine-protein phosphatase PP1-alpha catalytic subunit monomer, 38 kDa Homo sapiens protein
Apoptosis-stimulating of p53 protein 2 monomer, 26 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris, 150 mM NaCl, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 May 31
|
Flexible Tethering of ASPP Proteins Facilitates PP-1c Catalysis.
Structure 27(10):1485-1496.e4 (2019)
Zhou Y, Millott R, Kim HJ, Peng S, Edwards RA, Skene-Arnold T, Hammel M, Lees-Miller SP, Tainer JA, Holmes CFB, Glover JNM
|
| RgGuinier |
2.9 |
nm |
| Dmax |
10.2 |
nm |
| VolumePorod |
144 |
nm3 |
|
|
|
|
|
|
|
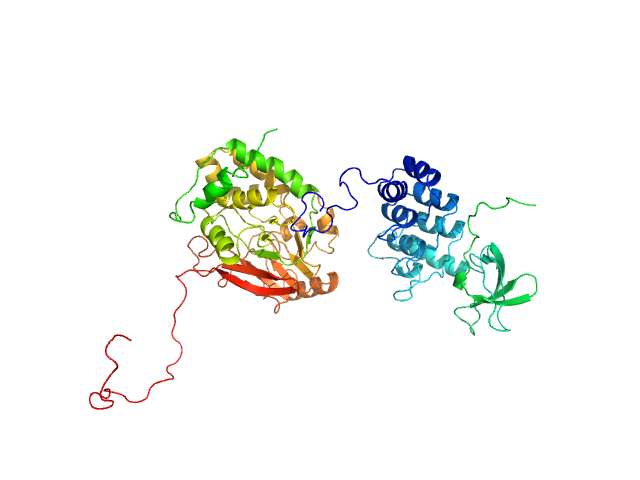
| Sample: |
Serine/threonine-protein phosphatase PP1-alpha catalytic subunit monomer, 38 kDa Homo sapiens protein
Apoptosis-stimulating of p53 protein 2 monomer, 26 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris, 150 mM NaCl, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 May 31
|
Flexible Tethering of ASPP Proteins Facilitates PP-1c Catalysis.
Structure 27(10):1485-1496.e4 (2019)
Zhou Y, Millott R, Kim HJ, Peng S, Edwards RA, Skene-Arnold T, Hammel M, Lees-Miller SP, Tainer JA, Holmes CFB, Glover JNM
|
| RgGuinier |
3.3 |
nm |
| Dmax |
11.7 |
nm |
| VolumePorod |
172 |
nm3 |
|
|