|
|
|
|
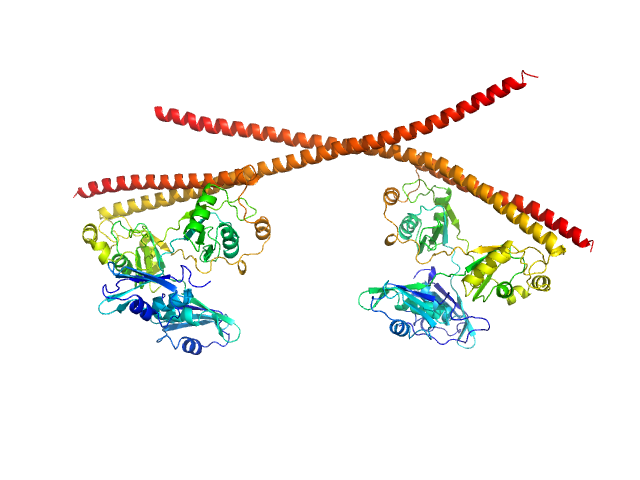
|
| Sample: |
Splicing factor, proline- and glutamine-rich dimer, 153 kDa Homo sapiens protein
Non-POU domain-containing octamer-binding protein dimer, 60 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris pH 7.5, 250 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Jun 18
|
Structural plasticity of the coiled-coil interactions in human SFPQ.
Nucleic Acids Res (2024)
Koning HJ, Lai JY, Marshall AC, Stroeher E, Monahan G, Pullakhandam A, Knott GJ, Ryan TM, Fox AH, Whitten A, Lee M, Bond CS
|
| RgGuinier |
5.5 |
nm |
| Dmax |
20.4 |
nm |
| VolumePorod |
304 |
nm3 |
|
|
|
|
|
|
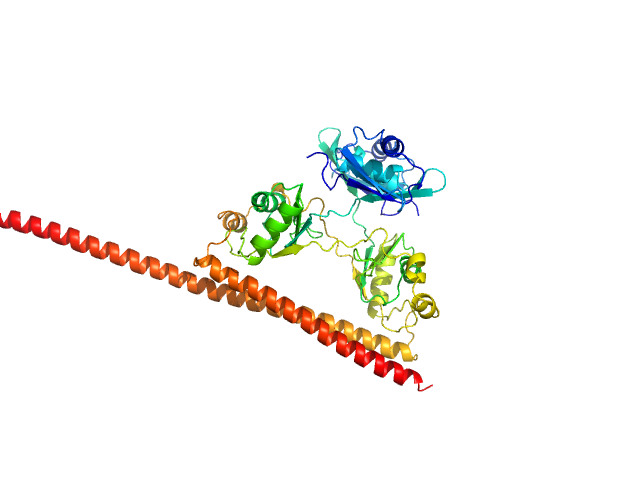
|
| Sample: |
Splicing factor, proline- and glutamine-rich (276-565) monomer, 34 kDa Homo sapiens protein
Non-POU domain-containing octamer-binding protein (53-312) monomer, 30 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris pH 7.5, 250 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2015 Apr 28
|
Structural plasticity of the coiled-coil interactions in human SFPQ.
Nucleic Acids Res (2024)
Koning HJ, Lai JY, Marshall AC, Stroeher E, Monahan G, Pullakhandam A, Knott GJ, Ryan TM, Fox AH, Whitten A, Lee M, Bond CS
|
| RgGuinier |
3.0 |
nm |
| Dmax |
12.7 |
nm |
| VolumePorod |
102 |
nm3 |
|
|
|
|
|
|
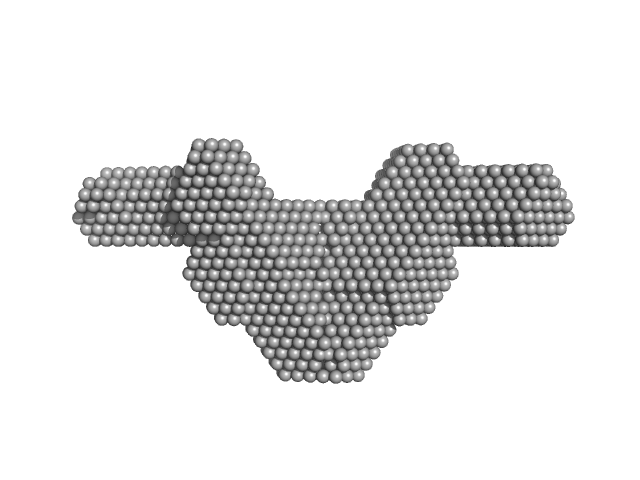
|
| Sample: |
Non-POU domain-containing octamer-binding protein dimer, 60 kDa Homo sapiens protein
Splicing factor, proline- and glutamine-rich SFPQ276-598(R542C)/NONO53-312 dimer) dimer, 76 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris pH 7.5, 250 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2015 Apr 28
|
Structural plasticity of the coiled-coil interactions in human SFPQ.
Nucleic Acids Res (2024)
Koning HJ, Lai JY, Marshall AC, Stroeher E, Monahan G, Pullakhandam A, Knott GJ, Ryan TM, Fox AH, Whitten A, Lee M, Bond CS
|
| RgGuinier |
5.2 |
nm |
| Dmax |
21.0 |
nm |
| VolumePorod |
257 |
nm3 |
|
|
|
|
|
|
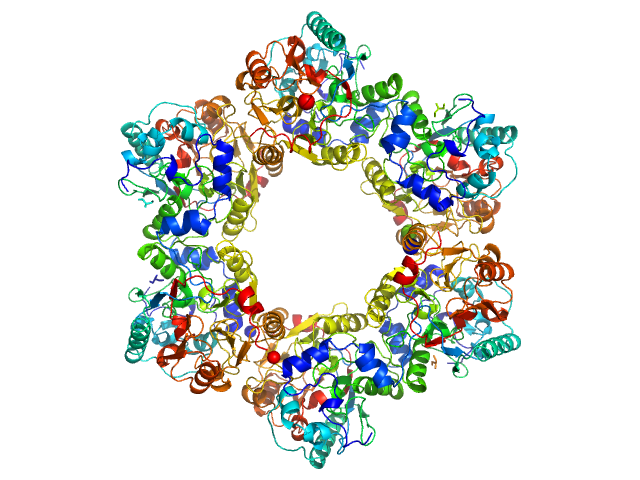
|
| Sample: |
2-nitroimidazole nitrohydrolase (T2I, G14D, K73R) hexamer, 258 kDa Mycobacterium sp. (strain … protein
|
| Buffer: |
25 mM Tris-HCl, 150 mM NaCl, 5% (v/v) glycerol, pH: 7.6 |
| Experiment: |
SAXS
data collected at BioSAXS, Australian Synchrotron on 2024 Aug 6
|
Structural insights into the enzymatic breakdown of azomycin-derived antibiotics by 2-nitroimdazole hydrolase (NnhA).
Commun Biol 7(1):1676 (2024)
Ahmed FH, Liu JW, Royan S, Warden AC, Esquirol L, Pandey G, Newman J, Scott C, Peat TS
|
| RgGuinier |
4.6 |
nm |
| Dmax |
12.6 |
nm |
| VolumePorod |
363 |
nm3 |
|
|
|
|
|
|
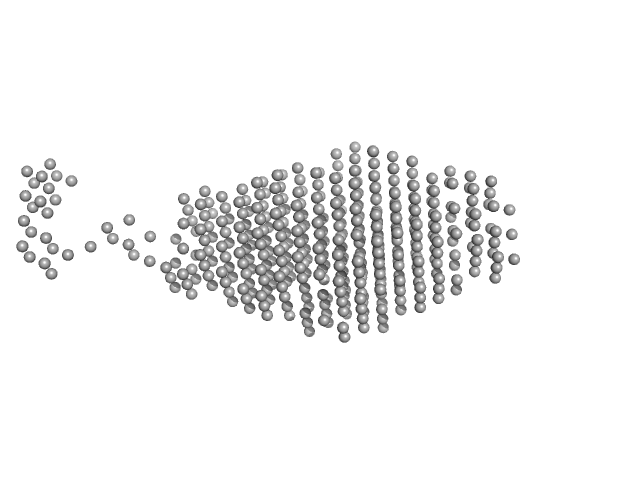
|
| Sample: |
Sensor domain-containing diguanylate cyclase dimer, 72 kDa Methylotenera sp. protein
|
| Buffer: |
10 mM Tris, 50 mM NaCl, 2 mM MgCl2, 3% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Apr 24
|
LOV-activated diguanylate cyclase
Ursula Vide
|
| RgGuinier |
3.4 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
128 |
nm3 |
|
|
|
|
|
|
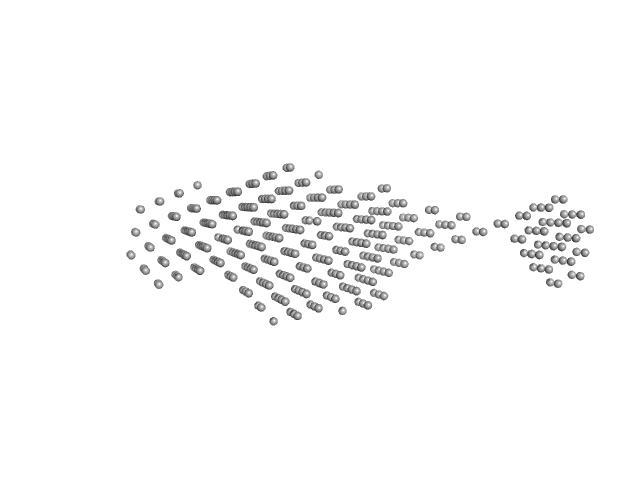
|
| Sample: |
Sensor domain-containing diguanylate cyclase dimer, 72 kDa Methylotenera sp. protein
|
| Buffer: |
10 mM Tris, 50 mM NaCl, 2 mM MgCl2, 3% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Apr 24
|
LOV-activated diguanylate cyclase
|
| RgGuinier |
5.2 |
nm |
| Dmax |
18.0 |
nm |
| VolumePorod |
159 |
nm3 |
|
|
|
|
|
|
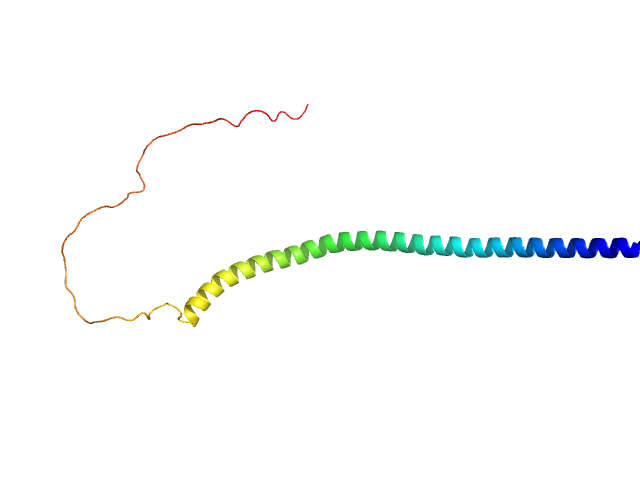
|
| Sample: |
Human alpha-synuclein monomer, 14 kDa Homo sapiens protein
|
| Buffer: |
10 mM HEPES, 50 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR - Institute of Microbial Technology (IMTech) on 2015 Mar 18
|
Visualizing gaussian-chain like structural models of human α-synuclein in monomeric pre-fibrillar state: Solution SAXS data and modeling analysis.
Int J Biol Macromol 288:138614 (2024)
Dey M, Gupta A, Badmalia MD, Ashish, Sharma D
|
| RgGuinier |
4.7 |
nm |
| Dmax |
17.6 |
nm |
| VolumePorod |
81 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Human alpha-synuclein monomer, 14 kDa Homo sapiens protein
|
| Buffer: |
10 mM HEPES, 50 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR - Institute of Microbial Technology (IMTech) on 2015 Mar 18
|
Visualizing gaussian-chain like structural models of human α-synuclein in monomeric pre-fibrillar state: Solution SAXS data and modeling analysis.
Int J Biol Macromol 288:138614 (2024)
Dey M, Gupta A, Badmalia MD, Ashish, Sharma D
|
| RgGuinier |
4.7 |
nm |
| Dmax |
18.0 |
nm |
| VolumePorod |
76 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Na/Ca-exchange protein, isoform D monomer, 31 kDa Drosophila melanogaster protein
|
| Buffer: |
20 mM TRIS, 200 mM NaCl, 1% v/v glycerol, 0.03% w/v NaN3, 1 mM β-mercaptoethanol, 2 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at Xenocs Xeuss, Institute of Physics, University of São Paulo on 2018 Mar 8
|
SAXS characterisation of CALX1.2 CBD12 construct
Roberto Kopke Salinas
|
| RgGuinier |
2.6 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
48 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Na/Ca-exchange protein, isoform D monomer, 31 kDa Drosophila melanogaster protein
|
| Buffer: |
20 mM TRIS, 200 mM NaCl, 1% v/v glycerol, 0.03% w/v NaN3, 1 mM β-mercaptoethanol, 50 mM CaCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at Xenocs Xeuss, Institute of Physics, University of São Paulo on 2018 Mar 8
|
SAXS characterisation of CALX1.2 CBD12 construct
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.9 |
nm |
| VolumePorod |
32 |
nm3 |
|
|