|
|
|
|
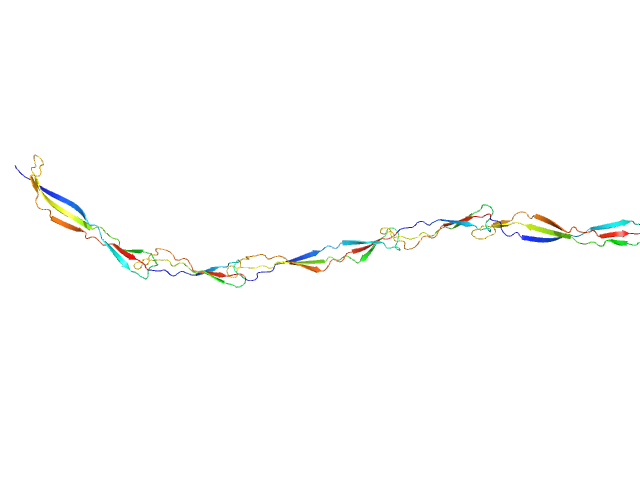
|
| Sample: |
Surface protein G monomer, 39 kDa Staphylococcus aureus protein
|
| Buffer: |
20 mM Tris 200 mM NaCl 1 mM EDTA 20 mM Tris.Cl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Nov 12
|
Cooperative folding of intrinsically disordered domains drives assembly of a strong elongated protein.
Nat Commun 6:7271 (2015)
Gruszka DT, Whelan F, Farrance OE, Fung HK, Paci E, Jeffries CM, Svergun DI, Baldock C, Baumann CG, Brockwell DJ, Potts JR, Clarke J
|
| RgGuinier |
7.7 |
nm |
| Dmax |
30.5 |
nm |
| VolumePorod |
49 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Surface protein G monomer, 53 kDa Staphylococcus aureus protein
|
| Buffer: |
20 mM Tris 200 mM NaCl 1 mM EDTA 20 mM Tris.Cl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Nov 12
|
Cooperative folding of intrinsically disordered domains drives assembly of a strong elongated protein.
Nat Commun 6:7271 (2015)
Gruszka DT, Whelan F, Farrance OE, Fung HK, Paci E, Jeffries CM, Svergun DI, Baldock C, Baumann CG, Brockwell DJ, Potts JR, Clarke J
|
| RgGuinier |
9.7 |
nm |
| Dmax |
38.5 |
nm |
| VolumePorod |
58 |
nm3 |
|
|
|
|
|
|
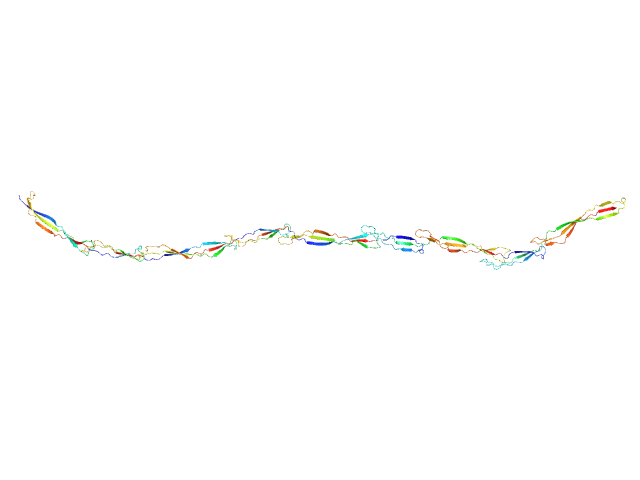
|
| Sample: |
Surface protein G monomer, 67 kDa Staphylococcus aureus protein
|
| Buffer: |
20 mM Tris 200 mM NaCl 1 mM EDTA 20 mM Tris.Cl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Nov 12
|
Cooperative folding of intrinsically disordered domains drives assembly of a strong elongated protein.
Nat Commun 6:7271 (2015)
Gruszka DT, Whelan F, Farrance OE, Fung HK, Paci E, Jeffries CM, Svergun DI, Baldock C, Baumann CG, Brockwell DJ, Potts JR, Clarke J
|
| RgGuinier |
12.0 |
nm |
| Dmax |
48.0 |
nm |
| VolumePorod |
87 |
nm3 |
|
|
|
|
|
|
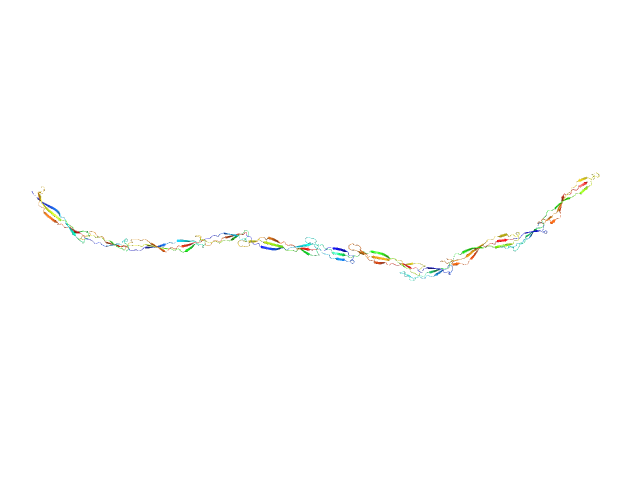
|
| Sample: |
Surface protein G monomer, 81 kDa Staphylococcus aureus protein
|
| Buffer: |
20 mM Tris 200 mM NaCl 1 mM EDTA 20 mM Tris.Cl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Nov 12
|
Cooperative folding of intrinsically disordered domains drives assembly of a strong elongated protein.
Nat Commun 6:7271 (2015)
Gruszka DT, Whelan F, Farrance OE, Fung HK, Paci E, Jeffries CM, Svergun DI, Baldock C, Baumann CG, Brockwell DJ, Potts JR, Clarke J
|
| RgGuinier |
14.1 |
nm |
| Dmax |
57.0 |
nm |
| VolumePorod |
89 |
nm3 |
|
|
|
|
|
|
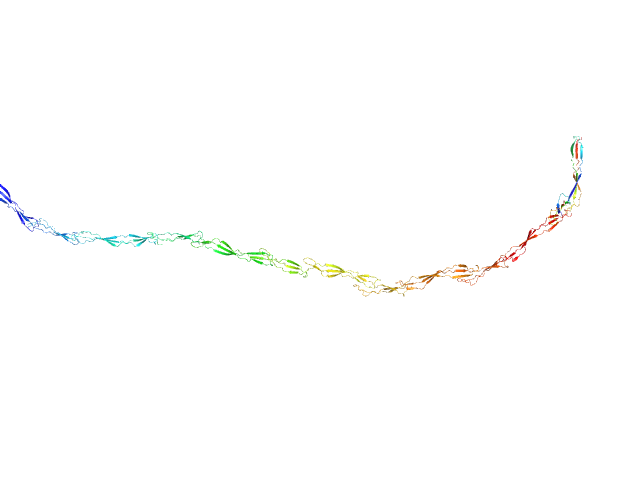
|
| Sample: |
Surface protein G monomer, 95 kDa Staphylococcus aureus protein
|
| Buffer: |
20 mM Tris 200 mM NaCl 1 mM EDTA 20 mM Tris.Cl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Nov 12
|
Cooperative folding of intrinsically disordered domains drives assembly of a strong elongated protein.
Nat Commun 6:7271 (2015)
Gruszka DT, Whelan F, Farrance OE, Fung HK, Paci E, Jeffries CM, Svergun DI, Baldock C, Baumann CG, Brockwell DJ, Potts JR, Clarke J
|
| RgGuinier |
15.9 |
nm |
| Dmax |
63.0 |
nm |
| VolumePorod |
122 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
PlaB tetramer, 220 kDa Legionella pneumophila protein
|
| Buffer: |
100 mM Tris 100 mM Nacl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Nov 15
|
Automated pipeline for purification, biophysical and x-ray analysis of biomacromolecular solutions.
Sci Rep 5:10734 (2015)
Graewert MA, Franke D, Jeffries CM, Blanchet CE, Ruskule D, Kuhle K, Flieger A, Schäfer B, Tartsch B, Meijers R, Svergun DI
|
| RgGuinier |
4.0 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
270 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Serine/threonine-protein kinase ATG1 dimer, 61 kDa Kluyveromyces lactis protein
Autophagy-related protein 13 dimer, 17 kDa Kluyveromyces lactis protein
Autophagy-related protein 17 dimer, 100 kDa Kluyveromyces lactis protein
Autophagy-related protein 29 dimer, 20 kDa Kluyveromyces lactis protein
KLLA0A10637p dimer, 32 kDa Kluyveromyces lactis protein
|
| Buffer: |
20 mM Tris, 200 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2013 Dec 20
|
Solution structure of the Atg1 complex: implications for the architecture of the phagophore assembly site.
Structure 23(5):809-818 (2015)
Köfinger J, Ragusa MJ, Lee IH, Hummer G, Hurley JH
|
| RgGuinier |
10.3 |
nm |
| Dmax |
34.0 |
nm |
| VolumePorod |
1000 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Serine/threonine-protein kinase ATG1 dimer, 61 kDa Kluyveromyces lactis protein
Autophagy-related protein 13 dimer, 17 kDa Kluyveromyces lactis protein
|
| Buffer: |
20 mM Tris, 200 mM NaCl, 2% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2013 Dec 10
|
Solution structure of the Atg1 complex: implications for the architecture of the phagophore assembly site.
Structure 23(5):809-818 (2015)
Köfinger J, Ragusa MJ, Lee IH, Hummer G, Hurley JH
|
| RgGuinier |
3.3 |
nm |
| Dmax |
10.9 |
nm |
| VolumePorod |
160 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Autophagy-related protein 17 dimer, 100 kDa Kluyveromyces lactis protein
Autophagy-related protein 29 dimer, 20 kDa Kluyveromyces lactis protein
KLLA0A10637p dimer, 32 kDa Kluyveromyces lactis protein
|
| Buffer: |
20 mM Tris, 200 mM NaCl, 2% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2013 Dec 10
|
Solution structure of the Atg1 complex: implications for the architecture of the phagophore assembly site.
Structure 23(5):809-818 (2015)
Köfinger J, Ragusa MJ, Lee IH, Hummer G, Hurley JH
|
| RgGuinier |
10.1 |
nm |
| Dmax |
34.0 |
nm |
| VolumePorod |
848 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Lysozyme C monomer, 14 kDa Gallus gallus protein
|
| Buffer: |
20 mM Sodium Acetate/HEPES, pH: 6.8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Feb 17
|
Correlation Map, a goodness-of-fit test for one-dimensional X-ray scattering spectra.
Nat Methods 12(5):419-22 (2015)
Franke D, Jeffries CM, Svergun DI
|
| RgGuinier |
1.5 |
nm |
| Dmax |
4.8 |
nm |
| VolumePorod |
24 |
nm3 |
|
|