|
|
|
|
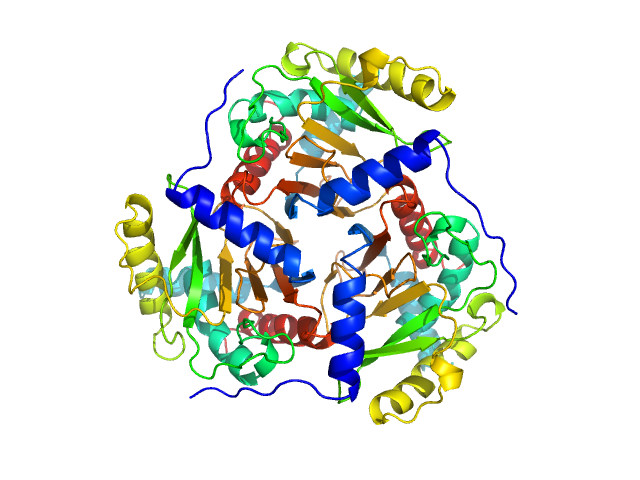
|
| Sample: |
Regulatory protein E2 trimer, 73 kDa Human papillomavirus type … protein
|
| Buffer: |
50 mM Tris ⁄ HCl, pH 8.8, 100 mM NaCl, pH: 8.8 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 May 27
|
Why are the 2-oxoacid dehydrogenase complexes so large? Generation of an active trimeric complex
Biochemical Journal 463(3):405-412 (2014)
Marrott N, Marshall J, Svergun D, Crennell S, Hough D, van den Elsen J, Danson M
|
| RgGuinier |
3.1 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
149 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Netrin-1 monomer, 49 kDa Homo sapiens protein
|
| Buffer: |
25 MES mM 200 mM NaCl 50 mM Tris 0.2 M ammonium sulfate (NH4)2(SO4) 1mM calcium chloride CaCl2, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Aug 26
|
The crystal structure of netrin-1 in complex with DCC reveals the bifunctionality of netrin-1 as a guidance cue.
Neuron 83(4):839-849 (2014)
Finci LI, Krüger N, Sun X, Zhang J, Chegkazi M, Wu Y, Schenk G, Mertens HDT, Svergun DI, Zhang Y, Wang JH, Meijers R
|
| RgGuinier |
3.9 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
100 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Deleted in Colorectal Cancer (FN5 & FN6) monomer, 26 kDa Homo sapiens protein
|
| Buffer: |
25 MES mM 200 mM NaCl 50 mM Tris 0.2 M ammonium sulfate (NH4)2(SO4) 1mM calcium chloride CaCl2, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Aug 26
|
The crystal structure of netrin-1 in complex with DCC reveals the bifunctionality of netrin-1 as a guidance cue.
Neuron 83(4):839-849 (2014)
Finci LI, Krüger N, Sun X, Zhang J, Chegkazi M, Wu Y, Schenk G, Mertens HDT, Svergun DI, Zhang Y, Wang JH, Meijers R
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
38 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Netrin-1 monomer, 49 kDa Homo sapiens protein
Deleted in Colorectal Cancer (FN5 & FN6) monomer, 26 kDa Homo sapiens protein
|
| Buffer: |
25 MES mM 200 mM NaCl 50 mM Tris 0.2 M ammonium sulfate (NH4)2(SO4) 1mM calcium chloride CaCl2, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Oct 31
|
The crystal structure of netrin-1 in complex with DCC reveals the bifunctionality of netrin-1 as a guidance cue.
Neuron 83(4):839-849 (2014)
Finci LI, Krüger N, Sun X, Zhang J, Chegkazi M, Wu Y, Schenk G, Mertens HDT, Svergun DI, Zhang Y, Wang JH, Meijers R
|
| RgGuinier |
5.1 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
120 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Netrin-1 monomer, 49 kDa Homo sapiens protein
Deleted in Colorectal Cancer (FN5 & FN6) M933R mutant monomer, 26 kDa Homo sapiens protein
|
| Buffer: |
25 MES mM 200 mM NaCl 50 mM Tris 0.2 M ammonium sulfate (NH4)2(SO4) 1mM calcium chloride CaCl2, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Aug 26
|
The crystal structure of netrin-1 in complex with DCC reveals the bifunctionality of netrin-1 as a guidance cue.
Neuron 83(4):839-849 (2014)
Finci LI, Krüger N, Sun X, Zhang J, Chegkazi M, Wu Y, Schenk G, Mertens HDT, Svergun DI, Zhang Y, Wang JH, Meijers R
|
| RgGuinier |
4.0 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
95 |
nm3 |
|
|
|
|
|
|
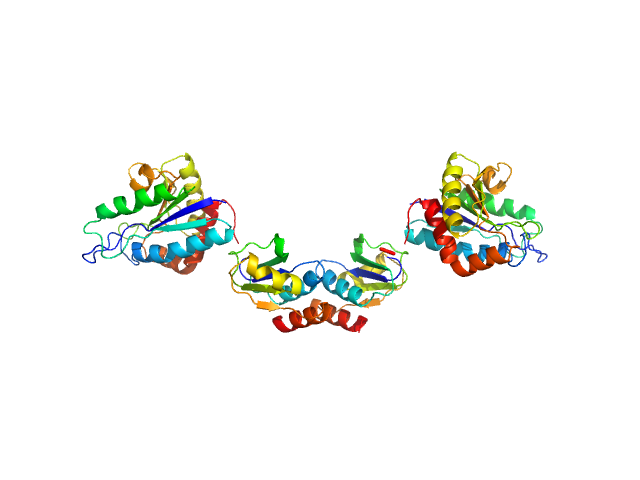
|
| Sample: |
Clostridium difficile bacteriophage 27 endolysin dimer, 64 kDa Clostridioides difficile protein
|
| Buffer: |
20 mM HEPES 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Mar 17
|
The CD27L and CTP1L endolysins targeting Clostridia contain a built-in trigger and release factor.
PLoS Pathog 10(7):e1004228 (2014)
Dunne M, Mertens HD, Garefalaki V, Jeffries CM, Thompson A, Lemke EA, Svergun DI, Mayer MJ, Narbad A, Meijers R
|
| RgGuinier |
3.3 |
nm |
| Dmax |
10.6 |
nm |
| VolumePorod |
72 |
nm3 |
|
|
|
|
|
|
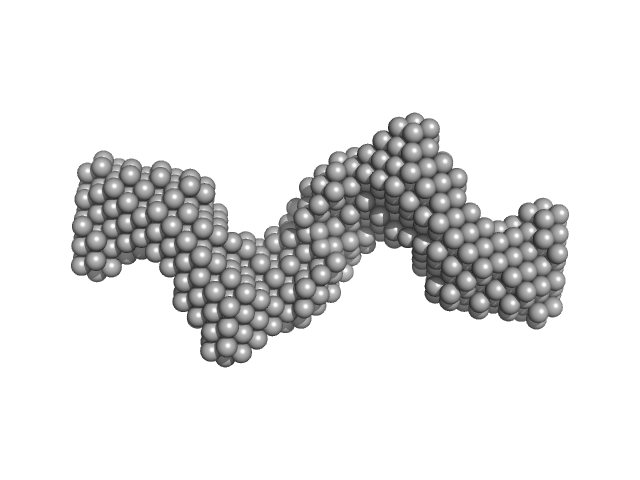
|
| Sample: |
Clostridium difficile bacteriophage 27 endolysin C238R mutant dimer, 64 kDa Clostridioides difficile protein
|
| Buffer: |
20 mM HEPES 500 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Jun 21
|
The CD27L and CTP1L endolysins targeting Clostridia contain a built-in trigger and release factor.
PLoS Pathog 10(7):e1004228 (2014)
Dunne M, Mertens HD, Garefalaki V, Jeffries CM, Thompson A, Lemke EA, Svergun DI, Mayer MJ, Narbad A, Meijers R
|
| RgGuinier |
4.2 |
nm |
| Dmax |
14.7 |
nm |
| VolumePorod |
92 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Ataxin-3 monomer, 41 kDa Homo sapiens protein
|
| Buffer: |
phosphate buffered saline (PBS), pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Dec 3
|
Interactions of ataxin-3 with its molecular partners in the protein machinery that sorts protein aggregates to the aggresome.
Int J Biochem Cell Biol 51:58-64 (2014)
Bonanomi M, Mazzucchelli S, D'Urzo A, Nardini M, Konarev PV, Invernizzi G, Svergun DI, Vanoni M, Regonesi ME, Tortora P
|
| RgGuinier |
6.9 |
nm |
| Dmax |
25.0 |
nm |
| VolumePorod |
425 |
nm3 |
|
|
|
|
|
|
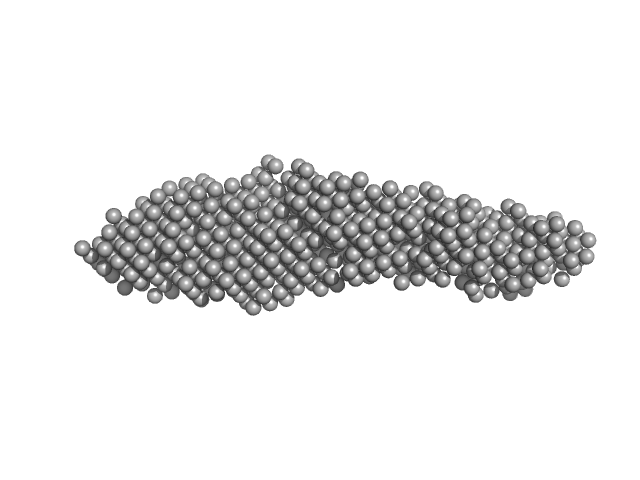
|
| Sample: |
Tubulin alpha-1A chain monomer, 50 kDa Homo sapiens protein
|
| Buffer: |
phosphate buffered saline (PBS), pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Dec 3
|
Interactions of ataxin-3 with its molecular partners in the protein machinery that sorts protein aggregates to the aggresome.
Int J Biochem Cell Biol 51:58-64 (2014)
Bonanomi M, Mazzucchelli S, D'Urzo A, Nardini M, Konarev PV, Invernizzi G, Svergun DI, Vanoni M, Regonesi ME, Tortora P
|
| RgGuinier |
7.0 |
nm |
| Dmax |
25.0 |
nm |
| VolumePorod |
340 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Ataxin-3 monomer, 41 kDa Homo sapiens protein
Tubulin alpha-1A chain monomer, 50 kDa Homo sapiens protein
|
| Buffer: |
phosphate buffered saline (PBS), pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Dec 3
|
Interactions of ataxin-3 with its molecular partners in the protein machinery that sorts protein aggregates to the aggresome.
Int J Biochem Cell Biol 51:58-64 (2014)
Bonanomi M, Mazzucchelli S, D'Urzo A, Nardini M, Konarev PV, Invernizzi G, Svergun DI, Vanoni M, Regonesi ME, Tortora P
|
| RgGuinier |
8.4 |
nm |
| Dmax |
30.0 |
nm |
| VolumePorod |
900 |
nm3 |
|
|